方案详情
文
与可见腐败和aprX表达水平有关的接种不同假单胞菌蛋白酶提取物的UHT牛奶的多肽图谱Peptidomic Fingerprints of Stored UHT Milk Inoculated with Protease Extracts from Different Pseudomonas Strains Relative to aprX Expression and Visible Spoilage
方案详情

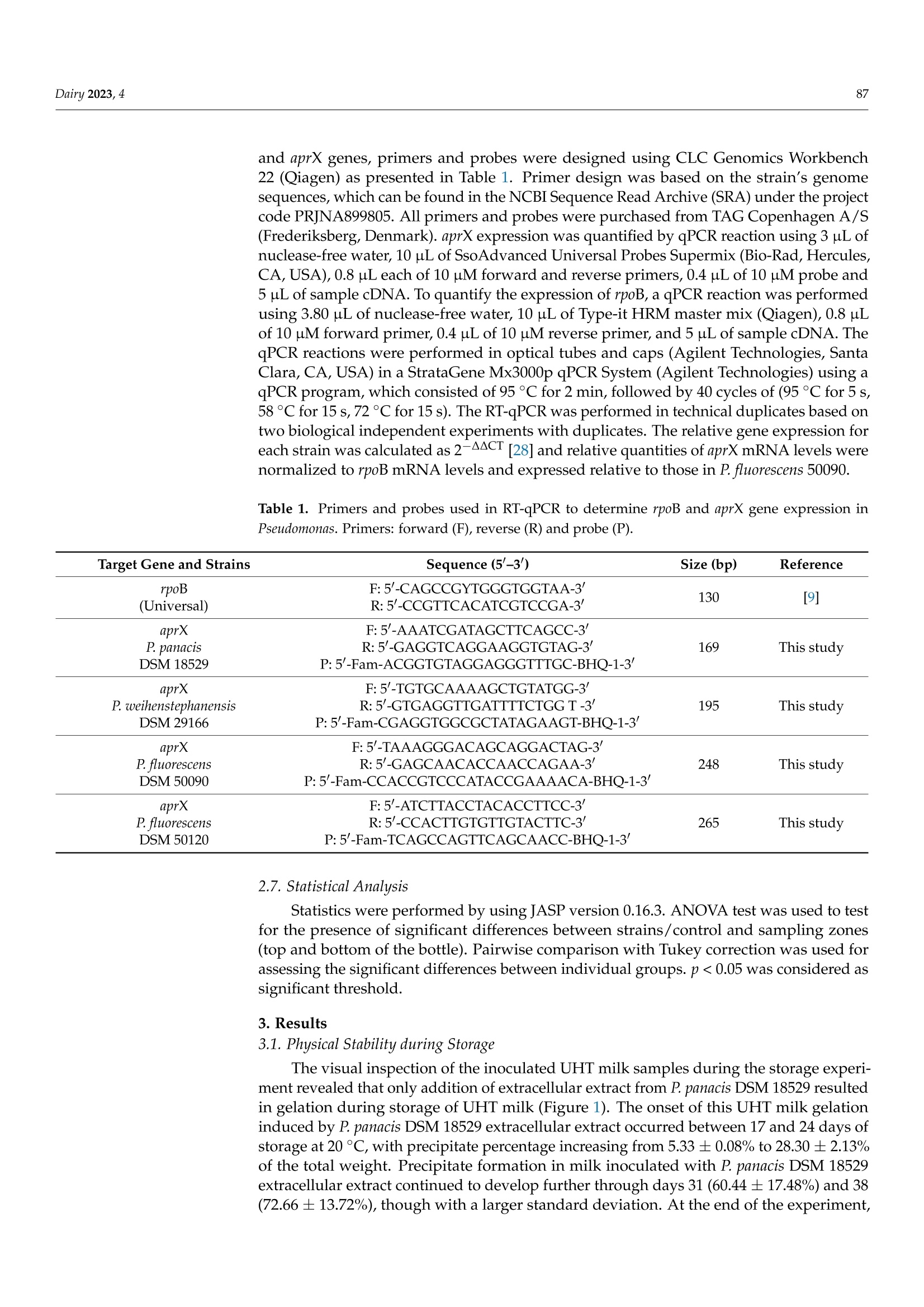
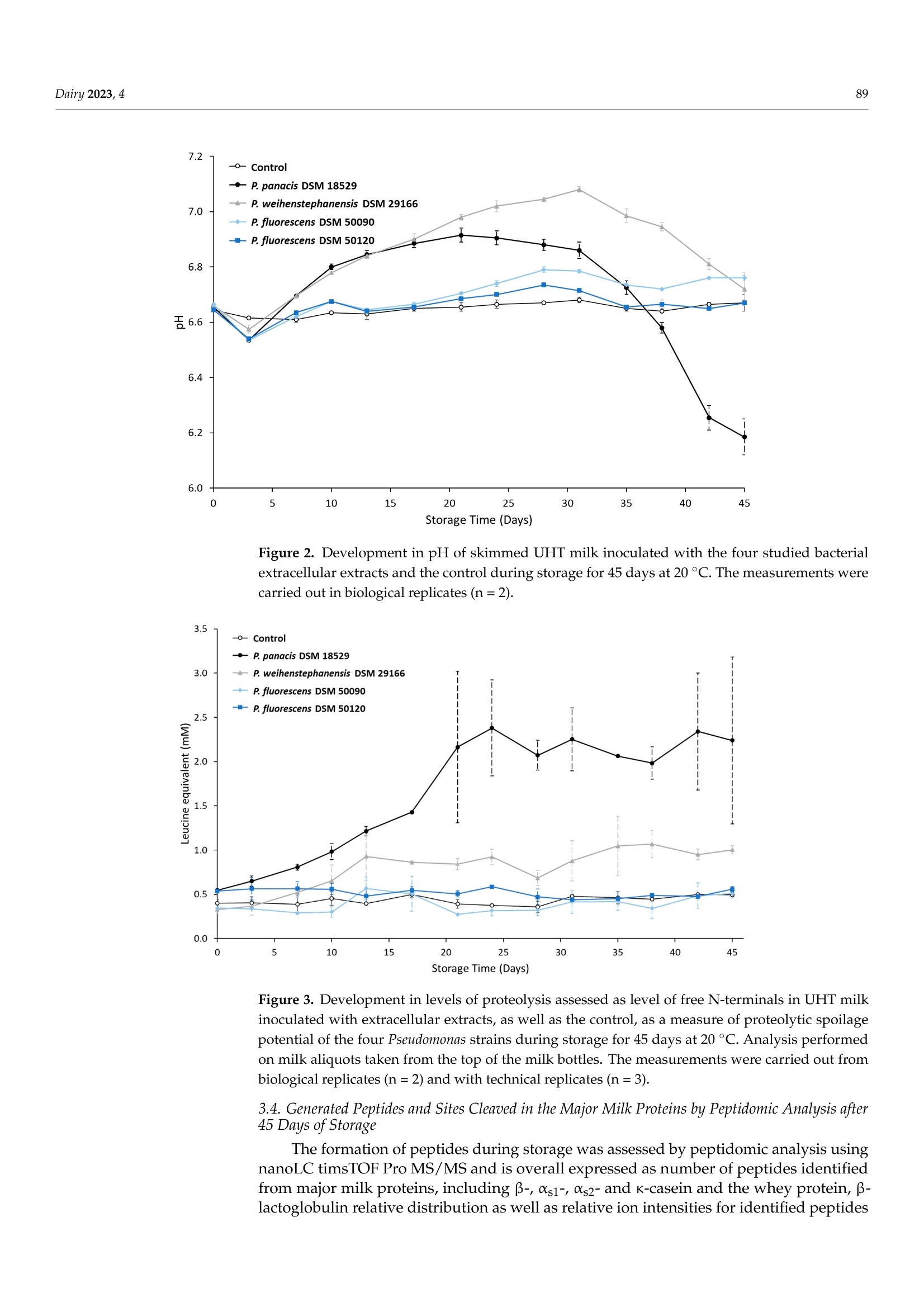
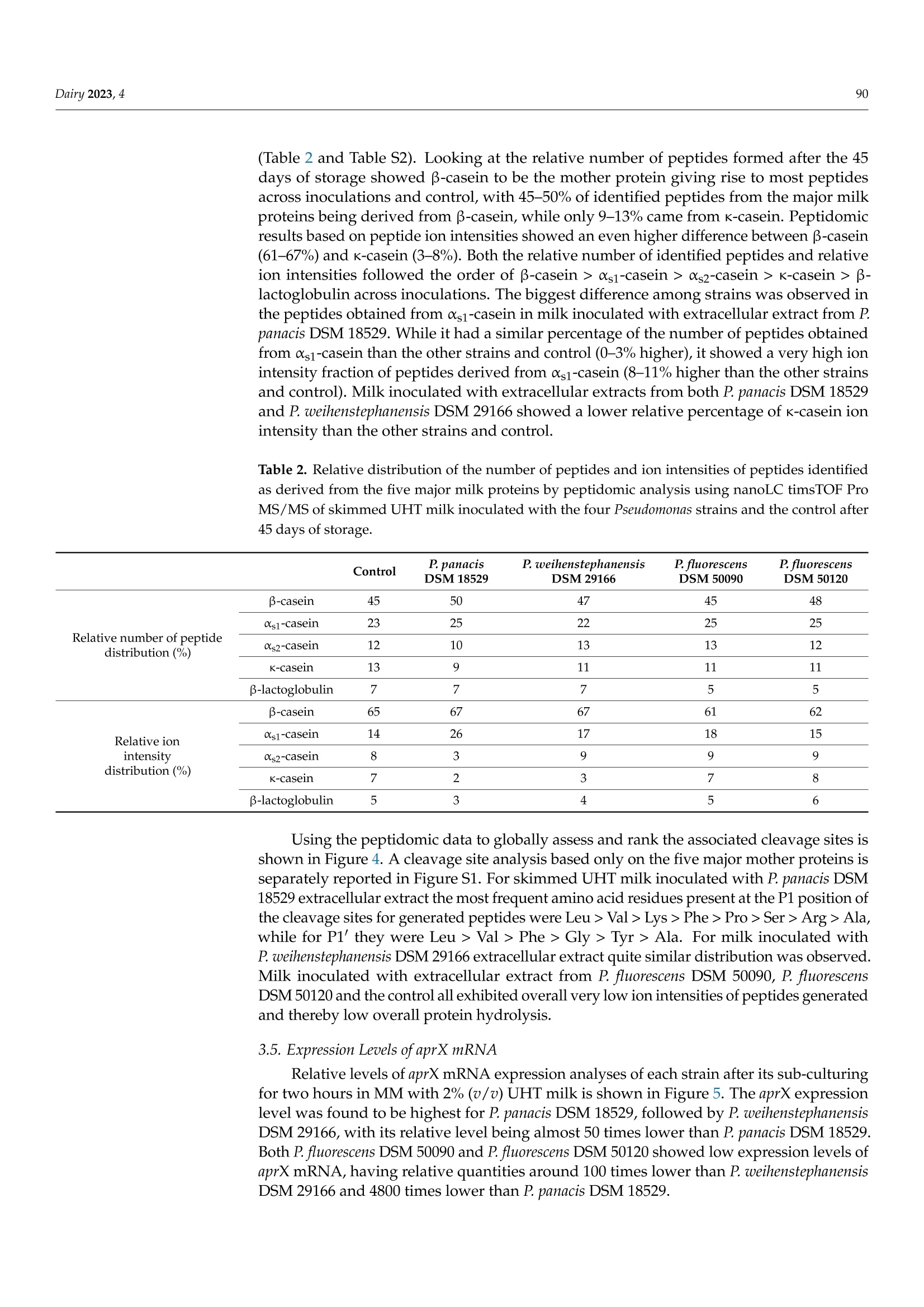
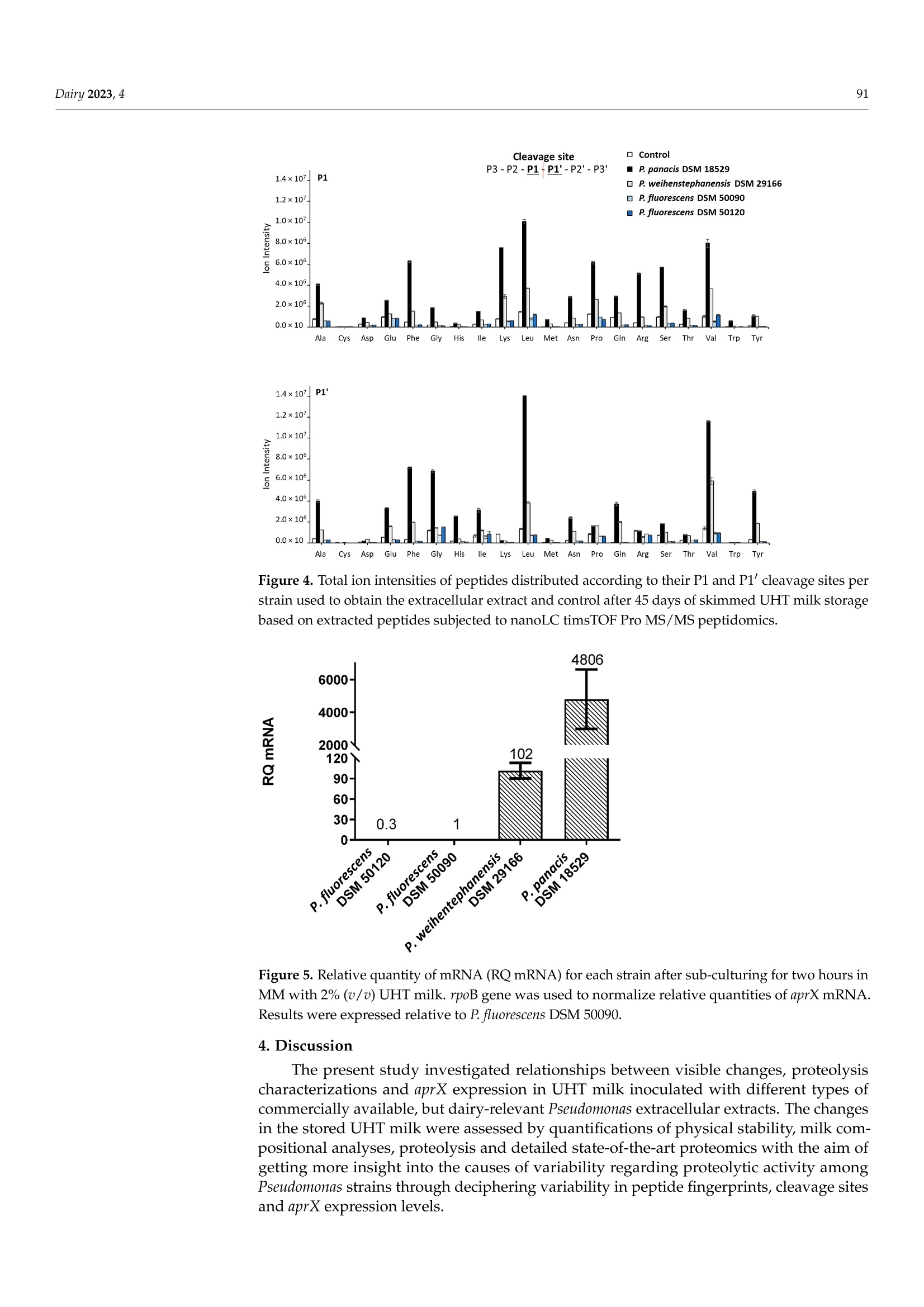
与可见腐败和aprX表达水平有关的接种不同假单胞菌蛋白酶提取物的UHT牛奶的多肽图谱Peptidomic Fingerprints of Stored UHT Milk Inoculated with Protease Extracts from Different Pseudomonas Strains Relative to aprX Expression and Visible Spoilage与可见腐败和aprX表达水平有关的接种不同假单胞菌蛋白酶提取物的UHT牛奶的多肽图谱 Dairy 2023, 484 Citation: Agui l era-Toro, M.;Nielsen,S.D.-H.; Kragh, M.L.;Xiao, Y.;Hansen, L .T .;Rauh, V.; Wiking, L.;Poulsen, N.A.; Larsen, L.B Peptidomic Fingerprints of Stored UHT Milk Inoculated with Protease Extracts from Different Pseudomonas Strains Relative to aprX Expression and Visible Spoilage. Dair y 2023,4,83-97. https://doi .org/10.3390/dairy4010005 Academic Editor: Hilton Deeth Received: 11 November 2022 Revised: 22 December 2022 Accepted: 27 December 2022 Published: 9 January 2023 Copyright: @ 2023 by the authors.L i censee MDPI, Basel, Switzerland.This article is an open access article distr i buted under the terms and conditions of the Creative Commons Attribution (CC BY ) l icense (https://creativecommons.org/licenses/by/4.0/) 2 Research Group for Food Microbiology and Hygiene, National Food Institute, Technica l University of Denmark, 2800 Kongens Lyngby , Denmark 3 Arla Innovation Center, Arla Foods Amba, 8200 Aarhus, Denmark Correspondence: miguel .agui l era@food.au.dk Abstract: Lately, concern about the protease AprX produced by Pseudomonas has increased in the dairy industry due to i ts ability to survive UHT treatment and spoi l UHT milk. Efficient prediction methods for UHT milk spoilage are currently lacking, mainly due to high diversity in proteolytic potential between Pseudomonas strains. The present study aimed to gain more insight into the variabi l ity between Pseudomonas strains regarding proteolytic potential by comparing thei r proteoly t ic capabil i ty wi t h t heir aprX expression l evels and differences in peptide formation. The variability in aprX expression levels in four Pseudomonas strains were related to physical stabi l ity, mi l k proteolysis and peptidomic cleavage patterns of milk proteins i n a storage experiment of UHT milk inoculated wi t h protease extracellular extracts and stored for 45 days at 20 C. A positive relationship was observed between the relat i ve expression of aprX and milk proteolysis during storage, with the strain Pseudomonas panacis DSM 18529 showing the highest level in both parameters. This strain was the only strain to show visual gelation, which occurred after 21 days. The peptide formation analysis showed a similar protein hydrolysis pattern between strains and high hydrolysis of as1-caseins during long-term spoilage putatively due to the activity of AprX was observed. Keywords: shelf-life; protein hydrolysis; peptide formation; gene expression 1. Introduction In recent years, the interest in ultra-high temperature (UHT) milk has increased due to i ts abi l ity to retain the quality during long-distance transport and storage under non-refrigerated conditions; which is key t o selling milk products at international markets, such as a growing Chinese dairy market [1,2]. UHT t reatment can eliminate microorganisms,but there are some bacterial spores and enzymes that can survive the heat t reatment,influencing the dairy product quality during storage. Among these, AprX, an alkaline zinc metalloprotease produced by Pseudomonas spp., has been pinpointed as one of the main causes of endogenous enzymatic spoilage in UHT milk [3-5]. In addition to its heat resistance, AprX relevance is due to the ability of Pseudomonas spp. to survive and grow in refrigerated milk, which makes this genus to most abundant in refrigerated raw milk [6]. AprX belongs to the serralysin family and has a molecular mass of 45-50 kDa [7]. The protease i s encoded by the aprX gene, which is l ocated in the aprX-lipA operon [8]. Current methods for spoilage prediction consist of the quantification of total bacterial count (TBC)and psychrotrophic bacteria counts (PBC), which are t ime consuming and unspecific for a correct assessment of the expected shelf-life prior to UHT processing [9]. New rapid meth-ods for spoilage prediction, such as qPCR and ELISA, have been suggested as alternatives for t he dairy i ndustry [10]. The successful implementation of an accurate and rapid method The aim of the present study was to gain more insight into the causes of proteolytic activity variabil i ty between Pseudomonas strains by comparing the spoilage potential of strains with their aprX expression variation and differences in peptide formation. UHT milk was inoculated with extracellular extracts from four commercially available strains of Pseudomonas and stored at 20 °C for 45 days. The strains comprised three dairy relevant and heat-resistant proteolytic enzyme producing strains, Pseudomonas panacis DSM 18529 [19],Pseudomonas weihenstephanensis DSM 29166 [20] and Pseudomonas fluorescens DSM 50120 [21];as well as Pseudomonas fluorescens DSM 50090 representing a low or even non-proteolytic strain, since i t has previously been reported to lack proteolytic activity [22]. Milk quality parameters i ncluding physical stability, pH and level of proteolysis were monitored during the storage experiment and related to aprX expression levels. Variability within specificities of proteases secreted by each strain towards milk proteins was deciphered by peptidomics. 2. Materials and Methods 2.1. Bacterial Strains and Preparation of Bacterial Extracellular Extracts The selected Pseudomonas strains (P. panacis DSM 18529, P. weihenstephanensis DSM 26166,P. f luorescens DSM 50090 and P. fluorescens DSM 50120) were obtained at the German Col-lection of Microorganisms and Cell Cultures (DSMZ) and frozen in glycerol at -80 °C for long-term storage. Strains were reactivated on tryptone soy agar (TSA, Oxoid, UK) plates for 24 h at 30 °C and grown in tryptic soy broth (TSB, Oxoid, UK) for 24 h at 30 °C. To quantify cell density, the TSB cultures were diluted i n maximum recovery di l uent (Oxoid,UK), spread plated on TSA and enumerated following overnight incubation of agar plates at 30 °C (Table S1). Preparation of bacterial extracellular extracts was carried out essentially as described by Zhang et al. [21]. To i nduce production of AprX, bacteria were harvested from 9mL of TSB cultures (24 h, 30 °C) by centrifugation (10 min, 20°C,4000× g) and transferred to 9 mL minimal media (MM) containing 2%(v/u) commercial UHT milk (3.5% fat), and subsequently cultured for 24 h at 25°C with gentle stirring. MM contained 2 g/L KH2PO4,7 g/L K2HPO4, 0.2 g/L MgSO4·7H20,2g/L (NH4)2SO4 and 8 g/L glycerol in MilliQ water. Bacterial cells were then removed by centrifugation (30 min, 4°℃, 10,800× g)and the supernatants containing the secreted extracellular proteases were recovered and concentrated by spin filtration (20 min, 4°℃,4000× g) using Amicon Ultra Filters (10 kDa cut-off, Millipore). These concentrates were then dialyzed against 10 mM potassium phosphate buffer, pH 7, for 48 h using Spectra/Por@3 Dialysis Membranes (3.5 kD cut- off). The dialyzed supernatants were freeze dried (Cooling Trap pro Teflon, CoolSafeM Scanvac, Lynge, Denmark) and stored at -80 C until further use. A standard extracellular extract was made for each strain by dissolving this freeze-dried material in 5 mL MilliQ water. Protein concentrations of the dissolved extracts were then determined by Dumas (Dumatherm, Gerhardt, Konigswinter, Germany). 2.2. UHT Milk Preparation, Storage Experiment and Sampling As substrate for the UHT milk storage test, fat free milk was obtained from commercial semi-skimmed UHT milk (1.5% fat) after extra skimming steps by centrifugation (30 min,4°C,2600× g) and separation of the fat layer by pipetting. This centrifugation process was repeated three times to ensure complete fat removal . A volume of 100 mL of skimmed mi l k were transferred to a test glass bottle used for the storage experiment, from where pH mea-surements and milk aliquots for further analyses were drawn twice a week, corresponding to t =0,3,7, 10, 13,17,21,24,28,31,35,38, 42 and 45 days. In parallel, seven Falcon tubes containing each 10 mL of skimmed milk aliquots were prepared in order to enable weekly assessment of visual gelation and/or precipitate formation during the storage period at t=3,10,17,24,41, 38 and 45 days after inoculation. This set-up (one 100 mL glass bottle and seven 10 mL Falcon tubes) was carried out in duplicate for each of the four extracellular extracts and the control (no extract added). Extracellular extracts were added to the 100 mL and 10 mL milk aliquots in volumes corresponding to 200 ug of extract protein per mL of milk. This concentration was estimated in a previous pilot experiment to be the necessary amount to observe proteolytic activity in 1 month (results not shown). To inhibit bacterial growth,0.1%(w/v) sodium azide was added to all milk samples. Stora KS ge was carried out a t 20 °C for 45 days. Mi l k pH was measured twice per week using a pH meter (PHM 92,Radiometer Copenhagen) by inserting t he probe directly into the 100 mL bottle without stirring. Additionally, twice a week, a milk aliquot was drawn from the top and the bottom regions of the 100 mL bottle. These aliquots were stored at -20 °C until further analyses. 2.3. Determination of Physical Stability in Inoculated UHT Milk during Storage 2.4. Determination of Proteolytic Act i vity i n Inoculated UHT Milk during Storage Proteolytic activity was measured by the fluorescamine assay, as described previ-ously [25], on aliquots taken from the storage experiment twice a week. Aliquots were mixed with an equal volume of 24% trichloroacetic acid (TCA), precipitated on ice for 30 min and centrifuged (20 min, 4°C,20,000× g). The supernatants were recovered and di-luted 1:2 with 1 mM HCl. Thirty-seven uL of each diluted supernatant was then mixed with 900 uL of 0.1 M Na-tetraborate and 300 uL of 0.2 mg/mL fluorescamine in dried acetone.Volumes of 250 uL from this mixture were transferred in triplicates to 96-microwell plates (white polystyrene microplates, Corning@, Numbrecht, Germany). Na-tetraborate (0.1 M)mixed with the fluorescamine solution was used as blank. Absorbance was measured by a spectrophotometer (multi-mode microplate reader SynergyTM 2, BioTeK, Winooski,VT , USA) using Ex 400 nm and Em 485 nm. Levels of free amino-terminals were expressed as L-leucine equivalents, calculated by the use of a standard curve based on leucine [25]. 2.5. Peptidomics of Inoculated UHT Milk from Storage Experiment Milk aliquots from day 0, 24 and 45 for each strain and control were used, and peptides were extracted from the UHT milk matrix according to Nielsen et al. [26]. Briefly the samples were added 20 mM triethylammonium bicarbonate (TEAB), reduced using 10 mM 1,4-dithioerythritiol and alkylated using 28 mM indole-3-acetic acid in 20 mM TEAB prior to precipitation of the proteins by adding ice-cold LC-MS grade acetonitrile (ACN).After centrifugation, the supernatants containing the peptides resulting from proteolysis were collected, lyophilized by Speedvac (Genevac EZ-2plus, BioLab, Ipswich, UK) at 35℃and diluted with 0.1% trifloroacetic acid. These resolubi l ized supernatants were desalted using PiercePeptide Desalting Spin Columns (Thermo Fisher Scientific, Waltham, MA,USA) according to the instructions of the manufacturer. Finally, the supernatants were lyophilized again, diluted with 2% ACN in 0.1% formic acid (FA), filtered with 10 kDa filters (Mini-Uniprep, Whatman, UK) and frozen at -20 °C until further analyzed. Peptide separation was performed on a nanoElute (Bruker, Solna, Sweden), with a captive-spray ionization source connected to a timsTOF PRO 2 (Bruker, Bremen, Germany).A volume of 10 uL of each resolubilized peptide extract was loaded onto a C18 PepSep 15 column (1.9×150 mm,inner diameter of 75 um, Bruker) at a column temperature of 50 °C . Peptides were eluted using 0.1% FA in water (solvent A) and 100% ACN, 0.1%FA (solvent B) with a flow rate of 400 nL/min. The 41 min gradient consisted of 2-17%solvent B for 22 min, 17-26% solvent B for 9 min, 26-37% solvent B for 5 min, 37-95%solvent B for 1 min and then 95% solvent B for 4 min. Spectra were collected in posi t ive ionization mode using data-dependent acquisition. The mass spectrometer was set to scan masses between 100 and 1700m/z. The collected spectra were analyzed by PEAKS software (Bioinformatics Solutions Inc, Waterloo, ON, Canada) as previously described [26]. Peptides were searched against a custom made database of 566 milk proteins identified in UHT milk when searching a smaller set of UHT milk samples against the total bovine proteome, including isoforms obtained from SwissProt, as well as common genetic variants of t he major mi l k proteins [26].Potential post-translational modifications included were carbamidomethylation (C), deami-dation (NQ), phosphorylation (STY), oxidation (M) and lactosylations (K). Analysis of amino acid residues present in the cleavage sites P1 and P1' was performed as described by Nielsen et al . [27], with P1 referring to the residue before a cleavage site and P1'referring to the residue after a cleavage site. 2.6. aprX mRNA Gene Expression For the aprX mRNA gene expression analysis, TSB cultures of the four strains were prepared as described in Section 2.1. Bacteria pellets were separated by centrifugation (10 min, 20°C, 4000×g) and resuspended in 5 mL of MM. A 1 mL aliquot of this solution was added to 9 mL of MM with 2% (v/v) UHT milk. Samples (n = 2 for each strain) were then incubated for 2 h at 25 °C with gentle stirring at 160 rpm. After incubation, a volume of 1.5 mL from each sample was added to pre-cooled (-20C) microcentrifuge tubes and centrifuged for 2 min at 9900× g. The supernatant was removed by decantation. Cell pellets were stored frozen at -20 °C unti l RNA extraction was performed on the following day using RNeasy Mini Kit (Qiagen, Sollentuna, Sweden) for total RNA isolation. Cell pellets were lyzed according to the manufacturer’s protocol with 600 uL RLT buffer (Qiagen,Sollentuna, Sweden) and 6 uL B-mercaptoethanol (Sigma-Aldrich, St. Louis, MO, USA)with two times mechanical disruption for 45 s (5 min break in between) (TissueLyser II,Qiagen) using 300 mg glass beads. The RNA concentration was quantified using Qubit 3.0 (Invitrogen, Carlsbad, CA, USA) with the Qubit RNA HS Assay Kit (Invitrogen). RNA samples were purified using TURBOTM DNase kit (2U/uL) (Ambion, Life Technologies,Naerum, Denmark) to remove contaminating gDNA. Sample RNA was reverse-transcribed into cDNA using the High Capacity cDNA Reverse Transcription Kit (Life Technologies,Naerum, Denmark) with the following program: 25 ℃ for 10 min, 37 °℃ for 120 min,85 °C for 5 min, and a cooling step to 4C. To quantify the mRNA levels of the rpoB and aprX genes, primers and probes were designed using CLC Genomics Workbench 22 (Qiagen) as presented in Table 1. Primer design was based on the strain’s genome sequences, which can be found in the NCBI Sequence Read Archive (SRA) under the project code PRJNA899805. All primers and probes were purchased from TAG Copenhagen A/S (Frederiksberg,Denmark). aprX expression was quantified by qPCR reaction using 3 uL of nuclease-free water, 10 uL of SsoAdvanced Universal Probes Supermix (Bio-Rad, Hercules,CA,USA), 0.8 uL each of 10 uM forward and reverse primers, 0.4 uL of 10 uM probe and 5 uL of sample cDNA. To quantify the expression of rpoB, a qPCR reaction was performed using 3.80 uL of nuclease-free water, 10 uL of Type-it HRM master mix (Qiagen), 0.8 uL of 10 uM forward primer, 0.4 uL of 10 uM reverse primer, and 5 uL of sample cDNA. The qPCR reactions were performed in optical tubes and caps (Agilent Technologies, Santa Clara, CA, USA) in a StrataGene Mx3000p qPCR System (Agilent Technologies ) using a qPCR program, which consisted of 95 °C for 2 min, followed by 40 cycles of (95°C for 5 s,58 ℃ for 15 s, 72 °C for 15 s). The RT-qPCR was performed i n technical dupl i cates based on two biological independent experiments with duplicates. The relative gene expression for each strain was calculated as 2-AAC T [28] and relative quantities of aprX mRNA levels were normalized to rpoB mRNA levels and expressed relative to those in P. fluorescens 50090. Table 1. Primers and probes used in RT-qPCR to determine rpoB and aprX gene expression i n Pseudomonas . Primers: forward (F), reverse (R) and probe (P). Target Gene and Strains Sequence (5'-3) Size (bp) Reference rpoB F: 5'-CAGCCGYTGGGTGGTAA-3' 130 [9] (Universal) R: 5'-CCGTTCACATCGTCCGA-3' aprX F:5'-AAATCGATAGCTTCAGCC-3' P. panacis R:5'-GAGGTCAGGAAGGTGTAG-3' 169 This study DSM 18529 P:5'-Fam-ACGGTGTAGGAGGGTTTGC-BHQ-1-3' aprX P. weihenstephanensis DSM 29166 F:5'-TGTGCAAAAGCTGTATGG-3' R: 5'-GTGAGGTTGATTTTCTGG T-3' P: 5-Fam-CGAGGTGGCGCTATAGAAGT-BHQ-1-3' 195 This study aprX P. fluorescens DSM 50090 F:5-TAAAGGGACAGCAGGACTAG-3' R:5'-GAGCAACACCAACCAGAA-3' P:5'-Fam-CCACCGTCCCATACCGAAAACA-BHQ-1-3' 248 This study 265 This study aprX F: 5'-ATCTTACCTACACCTTCC-3' P. fluorescens R:5'-CCACTTGTGTTGTACTTC-3' DSM 50120 P:5'-Fam-TCAGCCAGTTCAGCAACC-BHQ-1-3' 2.7. Statistical Analysis Statistics were performed by using JASP version 0.16.3. ANOVA test was used to test for the presence of significant differences between strains/control and sampling zones (top and bottom of the bottle). Pairwise comparison with Tukey correction was used for assessing the significant differences between individual groups. p <0.05 was considered as significant threshold. 3. Results 3.1. Physical Stability during Storage The visual inspection of the inoculated UHT milk samples during the storage experi-ment revealed that only addition of extracellular extract from P. panacis DSM 18529 resulted in gelation during storage of UHT milk (Figure 1). The onset of this UHT milk gelation induced by P. panacis DSM 18529 extracellular extract occurred between 17 and 24 days of storage a t 20 °C , with precipitate percentage increasing from 5.33 ±0.08% to 28.30±2.13%of the total weight. Precipitate formation in milk inoculated with P. panacis DSM 18529extracellular extract continued to develop further through days 31 (60.44±17.48%) and 38(72.66±13.72%), though with a l arger standard deviation. At the end of the experiment, on day 45, precipitate formation in UHT milk with added P. panacis DSM 18529 extract was determined to be 49.86 ±4.17%, thus almost half of the total sample weight. Precipitate formation by P. panacis DSM 18529 was significantly higher t han the other strains as well as the control (p<0.05). No signi f icant di f ferences in precipitate formation during storage were observed between the control and the UHT milk with added extracts from the other strains. The average precipitate formation in UHT mi l k i noculated with extracellular extract from P. weihenstephanensis DSM 29166, P. fluorescens DSM 50090, P. f luorescens DSM 50120and control during the storage experiment were 6.39 ±2.88%,5.21±2.32%,4.69±2.26%and 2.75±0.86%, respectively. Figure 1. Skimmed UHT milk inoculated with extracellular media from the four Pseudomonas strains and the control after 45 days of storage at 20 °C. From left to right, 1-2: control UHT milk,3-4: P. fluorescens DSM 50120, 5-6: P. weihenstephanensis DSM 29166, 7-8: P. fluorescens DSM 50090 and 9-10: P. panacis DSM 18529. 3.2. Development in pH during Storage Developments in pH of the stored, inoculated UHT skimmed milk and the control during storage is shown in Figure 2. The control and UHT milk inoculated with extracellular extract from P. fluorescens DSM 50120 and P. fluorescens DSM 50090 showed a stable pH over storage time. UHT milk inoculated with extracellular extract from P. panacis DSM 18529and P. weihenstephanensis DSM 29166 followed the same trend, where pH initially steadily increased over the f irst weeks and then decreased after 21 and 31 days, respectively. The UHT milk inoculated with P. panacis DSM 18529 extracellular extract reached a maximum pH value of 6.92 ±0.03 on day 21 and a minimum pH value of 6.19 ±0.07 on day 45. For the UHT milk inoculated with P. weihenstephanensis DSM 29166 extracellular extract, pH on day 0 was at its lowest with a pH value of 6.66 ±0.00 and reached its maximum on day 31, with pH value of 7.08 ±0.01. After 45 days, pH decreased back to pH 6.72 ±0.06.The average pH of control and UHT milk inoculated with extracellular extracts from P. fluorescens DSM 50090 and P. fluorescens DSM 50120 was 6.64 ±0.02,6.70±0.07 and 6.66 ±0.04, respectively. 3.3. Development in Proteolysis Level during Storage The development in the level of free N-terminals as a result of proteolysis in skimmed UHT milk samples during storage can be observed in Figure 3. UHT milk inoculated with extracel l ular extract from P. panacis DSM 18529 had the highest overall proteolytic ac t ivity (p <0.05) as well as the highest increase in level of proteolysis degree over storage time,reaching high levels from 21 days onwards with a l eucine equivalent of 2.16 ±0.86mM.The UHT milk inoculated with P. weihenstephanensis DSM 29166 extracellular extract was the second most proteolytic and exhibited a proteolytic level that was significantly (p<0.05)higher than UHT milk inoculated with extracellular extract from P. fluorescens DSM 50090,P. f luorescens DSM 50120 and the control. There was no significant di f ference in the prote-olytic activity between samples taken from the top or bottom layers of the bottles (results not shown). Storage Time (Days) Figure 2. Development in pH of skimmed UHT milk inoculated with the four studied bacterial extracel l ular extracts and the control during storage for 45 days at 20 °C. The measurements were carried out i n biological replicates (n=2). Figure 3. Development in levels of proteolysis assessed as level of free N-terminals in UHT milk inoculated with extracellular extracts, as well as the control, as a measure of proteolytic spoilage potentia l of the four Pseudomonas strains during storage for 45 days at 20 °C. Analysis performed on milk aliquots taken from the top of the milk bottles. The measurements were carried out from biological replicates (n=2) and with technica l replicates (n=3). 3.4. Generated Peptides and Sites Cleaved in the Major Milk Proteins by Peptidomic Analysis after 45 Days of Storage The formation of peptides during storage was assessed by peptidomic analysis using nanoLC timsTOF Pro MS/MS and is overal l expressed as number of peptides identified from major milk proteins, including B-, αs1-,心s2-and k-casein and the whey protein, β-lactoglobulin relative distribution as well as relative ion i ntensities for identified peptides (Table 2 and Table S2). Looking at the relative number of peptides formed after the 45days of storage showed B-casein to be the mother protein giving rise to most peptides across inoculations and control, with 45-50% of identi f ied peptides from the major milk proteins being derived from B-casein, while only 9-13% came from k-casein. Peptidomic results based on peptide ion intensities showed an even higher difference between B-casein (61-67%) and k-casein (3-8%). Both the relative number of identified peptides and relative ion intensi t ies followed the order of B-casein > αs1-casein> 心s2-casein > K-casein>β-lactoglobulin across inoculations. The biggest difference among strains was observed in the peptides obtained from &s1-casein in milk inoculated with extracellular extract from P.panacis DSM 18529. While it had a similar percentage of the number of peptides obtained from &s1-casein than the other strains and control (0-3% higher), it showed a very high ion intensity fraction of peptides derived from &s1-casein (8-11% higher than the other strains and control). Milk inoculated with extracellular extracts from both P. panacis DSM 18529and P. weihenstephanensis DSM 29166 showed a lower relative percentage of k-casein ion intensity than the other strains and control . Table 2. Relative distribution of the number of peptides and ion intensities of peptides ident i fied as derived from the f ive major milk proteins by peptidomic analysis using nanoLC timsTOF Pro MS/MS of skimmed UHT milk inoculated with the four Pseudomonas strains and the control after 45 days of storage. Control P. panacis DSM 18529 P. weihenstephanensis DSM 29166 P. fluorescens DSM 50090 P. fluorescens DSM 50120 Relative number of peptide distribution (%) B-casein 45 50 47 45 48 Xs1-casein 23 25 22 25 25 Xs2-casein 12 10 13 13 12 K-casein 13 9 11 11 11 Relative ion intensity distribution(%) B-lactoglobulin 7 7 7 5 5 B-casein 65 67 67 61 62 Xs1-casein 14 26 17 18 15 Xs2-casein 8 3 9 9 9 K-casein 7 2 3 7 8 β-lactoglobulin 5 3 4 5 6 Using t he peptidomic data to globally assess and rank the associated cleavage sites is shown in Figure 4. A cleavage site analysis based only on the f ive major mother proteins is separately reported in Figure S1. For skimmed UHT milk inoculated with P. panacis DSM 18529 extracellular extract the most frequent amino acid residues present at the P1 position of the cleavage si t es for generated peptides were Leu> Val> Lys>Phe> Pro > Ser> Arg>Ala,while for P1' they were Leu > Val> Phe > Gly > Tyr> Ala. For mi l k inoculated with P. weihenstephanensis DSM 29166 extracel l ular extract quite simi l ar distribution was observed.Milk inoculated with extracellular extract from P. fluorescens DSM 50090, P. fluorescens DSM 50120 and the control all exhibited overall very low ion intensities of peptides generated and thereby low overall protein hydrolysis. 3.5. Expression Levels ofaprXmRNA Relative level s of aprX mRNA expression analyses of each strain after i ts sub-culturing for two hours in MM with 2%(v/u) UHT milk is shown in Figure 5. The aprX expression level was found to be highest for P. panacis DSM 18529, followed by P. weihenstephanensis DSM 29166, with its relative l evel being almost 50 t i mes lower than P. panacis DSM 18529.Both P. fluorescens DSM 50090 and P. fluorescens DSM 50120 showed low expression levels of aprX mRNA, having relative quantities around 100 times lower t han P. weihenstephanensis DSM 29166 and 4800 times lower than P. panacis DSM 18529. Figure 4. Total ion intensities of peptides distributed according to their P1 and P1' cleavage sites per strain used to obtain the extracellular extract and control after 45 days of skimmed UHT milk storage based on extracted peptides subjected to nanoLC timsTOF Pro MS/MS peptidomics. Figure 5. Relative quantity of mRNA (RQmRNA) for each strain after sub-culturing for two hours in MM with 2%(u/u) UHT milk. rpoB gene was used to normalize relative quantities of aprX mRNA.Results were expressed relative to P. f luorescens DSM 50090. 4. Discussion The present study investigated relationships between visible changes, proteolysis characterizations and aprX expression in UHT milk inoculated with different types of commercially available, but dairy-relevant Pseudomonas extracellular extrac t s. The changes in t he stored UHT milk were assessed by quantifications of physical stabi l ity, milk com-posi t ional analyses, proteolysis and detailed state-of-the-art proteomics with the aim of getting more i nsight i nto the causes of variability regarding proteolytic activity among Pseudomonas strains through deciphering variability in peptide fingerprints, cleavage sites and aprX expression levels. 4.1. Relationship between Spoilage Potential, Milk Proteolysis and aprX Expression Milk proteolysis, determined as level of free N-terminals generated in stored UHT milk inoculated with extracellular extracts from Pseudomonas strains, was observed to comply with the relative aprX expression levels measured from the same Pseudomonas strains cultured in MM with 2%(v/v) UHT mi l k. UHT milk inoculated with extracellular extract from P. panacis DSM 18529, which showed the highest aprX expression levels, was found to have the highest proteolytic activity and aprX expression based on both fluorescamine assay and peptidomic results. The same observation was true for P. weihenstephanensis DSM 29166as the second highest proteolytic strain, having the second highest aprX expression levels.The two additional studied strains, P. fluorescens DSM 50090 and P. fluorescens DSM 50120showed low expression of aprX and low proteolytic activity. This is in accordance with previous reports of protease expression being the main driver of milk proteolysis, with individual differences between proteases having a minor impact [12]. 4.2. pH Changes and UHT Milk Spoilage during Storage 4.3. Peptidomics: Identified Peptides and Cleavage Sites Based on the information from both relative number of identified peptides and their relative ion intensities, protein hydrolysis by proteases secreted by the Pseudomonas strains was observed to follow the pattern B-casein > Cs1-casein > 仑s2-casein > K-casein > β-lactoglobulin. This order was observed for all strains, including P. panacis DSM 18529.These results are in accordance with the findings from other studies where peptide numbers were determined by LC-MS in UHT milk inoculated with either Pseudomonas cultures [14]or purified AprX [39]. This peptide hydrolysis pattern reflects milk protein composition,as β-and xs1-caseins represent the highest percentage of caseins, being around 35% for B-casein and 37% for &s1-casein, while xs2-casein accounts for 10% and K-casein for 12%in bovine milk, generally speaking [6]. By analysis of number of peptides formed by extracellular extracts from Pseudomonas inoculated on casein isolates, Mateos et al. [16]observed comparable results, with number of peptides following the order of B-casein> αs1-casein > k-casein. Stuknyte et al. [40], however, identified no major differences between the amount of peptides identified from B-casein, αs1-casein and k-casein when analyzing the number of peptides coming from different types of casein isolates. Zhang et al. [21] analyzed the casein hydrolysis in AprX-inoculated milk by RP-HPLC, where k-casein was observed to be t he f irst casein to be hydrolyzed (95% of k-casein hydrolyzed at gelation onset).In the present study, the major difference regarding peptide distribution between UHT milk inoculated with extracellular extract from P. panacis DSM 18529 and the other strains was a higher relative peptide ion intensity percentage derived from as1-casein in UHT milk inoculated with extracellular extract from P. panacis DSM 18529. In addi t ion, a lower relative peptide ion intensity percentage derive f rom k-casein was observed in UHT milk inoculated with extracellular extract from P. panacis DSM 18529 and P. weihenstephanensis DSM 291666. This could be explained by a preferred cleavage of the caseinomacropeptide region of k-casein, producing a higher quantity of glycosylated peptides, which are more difficul t to detect when using mass spectrometry [10]. These f indings suggest that, while k-casein hydrolysis might be the main target for AprX at the onset of storage, long-term exposure to the protease, with concomitant destabi l ization of the casein micelles, may lead to more prominent cleavage of as1-casein. Some researchers have compared the proteolytic effect of AprX with chymosin [10,41],which has a high specificity for the peptide bond Phe105-Met106 of k-casein. In addition,Reid et al. [42] and Garcia-Risco et al. [43] reported the presence of para -K-casein in milk of spoiled UHT milk samples. However, in Figure 4, it can be observed that, while there is a relatively high preference for Phe residues at the P1 position of cleavage sites for UHT milk inoculated with extracel l ular extracts from P. panacis DSM 18529 and P. weihenstephanensis DSM 291666, the preference for Met in P1' is rather low. Individual analysis of preferred cleavage sites in k-casein (Figure S1), showed for UHT milk inoculated with extracellular extract from P. panacis DSM 18529 a small preference for Phe and Met residues in P1 and P1'positions, respectively; however, with low ion intensities of the l inked peptides. Datta and Deeth [13] have previously discussed that AprX specificity for the Phe105-Met106bond is lower t han that of chymosin, while Mateos et al. [16] have reported AprX to have generally low cleavage site specificity. Mateos et al. [16] reported that the presence of basic (Arg, Lys, His) or aromatic (Tyr, Phe, Trp) amino acids in P1 positions was correlated with a higher susceptibility for cleavage of the bond by AprX, wh nr SuSC ile the presence of acidic (Asp,Glu, Ser) or aliphatic (Val, Ile, Pro) amino acids was unfavorable. For the P1'position, it was reported that presence of Va l , Met, Phe, Tyr, His and Gln was favorable, while Pro, Trp, Asp,Ser, Lys and Arg residues were unfavorable. In the present study, preferred P1 residues for cleavage varied in UHT milk inoculate with extracellular extracts from both P. panacis DSM 18529 and P. weihenstephanensis DSM 29166, being the two strains that exhibited milk proteolysis, from those described by Mateos et al. [16]. Only some of the basic (Arg, Lys)and aromatic (Phe) amino acids showed high ion i ntensity, while some of the reported unfavorable amino acids by Mateos et al.[16] (Ser, Val, Pro) were also identified with high intensity. The high amount of Lys and Arg residues in the P1 cleavage site for peptides produced by P. panacis DSM 18529 extracellular extracts, which are mainly products of αs1~and B-caseins (Figure S1), could also be explained by plasmin activity, since plasmin is highly speci f ic for cleavage after Lys and Arg. Fajardo-L i ra et al. [44] described an increased plasmin activity in the presence of bacterial proteases as a result of plasmin release from the casein micelles. On the other hand, Frohbieter et al . [45] concluded that bacterial proteases i nstead had a direct stimulatory effect on the plasminogen activators. Regarding P1', the observed results in this study for P. panacis DSM 18529 and P. weihenstephanensis DSM 291666 extracellular extracts matched the f indings from Mateos et al . [16], and may indicate a high importance of specific residues in the P1' site for AprX cleavage. 5. Conclusions Expression of AprX mRNA was found to have a strong relation with milk proteol-ysis, reinforcing the idea that variations in spoilage by AprX are driven by quantitative differences rather t han qualitative changes in the protein hydrolysis pattern. While previous studies have highlighted k-casein as a priority target for hydrolysis by AprX, the observed results showed that during long-term spoilage, αs1-caseins are the casein type that suffers the highest hydrolysis. P1' amino acids at the cleavage site for proteases, including AprX, in the extracellular media from Pseudomonas are broad but very conserved between studies, which may indicate a high impact of these cleavage sites for protein hydrolysis and spoilage potential. In addition, a cooperative effect seems to exist between bacterial proteases and plasmin, even though the specific mechanism is still not elucidated. Supplementary Materials: The following supporting information can be downloaded at: https://www.mdpi .com/art i cle/10.3390/dai ry 4010005/s1, Table S1: Bacterial growth of the four studied strains, Table S2: Number of identified peptides and ion intensity of the four studied strains and control, Figure S1: Ion intensity of peptides obtained from the five most common proteins (B-casein,Cs1-casein, αs2-casein, K-casein and β-lactoglobulin) distributed according to their P1 and P1'cleavage site for the four di f ferent strains and control after 45 days of storage. Author Contributions: Conceptualization, M.A.-T., L.B.L., N.A.P. and L.W.; methodology, M.A.-T.,S.D.-H.N., M.L .K., Y.X., V.R., L .B.L ., N.A.P. and L .T.H.; validation, formal analysis and investigation,M.A.-T., S.D.-H.N. and M.L.K.; data curation, M.A.-T., S.D.-H.N. and M.L.K.; writing-original draft preparation, M.A.-T ., L.B.L., N.A.P. and L.W.; wri t ing-review and editing, M.A.-T., S.D.-H.N.,M.L .K., Y.X., V.R.,L.B.L.,N.A.P., L.T.H. and L.W.; visualization, M.A.-T.; supervision, L.B.L., N.A.P.,L.W., Y.X. and L.T .H.; project administration, L.B.L.; funding acquisition, L.B.L. All authors have read and agreed to the published version of the manuscript. Funding: This research was founded by Milk Levy Fund , Danish Research Foundation, Arla Foods Amba, Aarhus University and Danish Technical University. Institutional Review Board Statement: Not applicable Informed Consent Statement: Not applicable. Data Availability Statement: Data is contained within the article or Supplementary Material. Acknowledgments: The authors would like to thank all the involved lab technicians at the Aarhus University, Danish Technica l University and Arla Innovation Center laboratories for thei r assistance with the performed analyses. Conflicts of Interest: The authors declare no conflict of interest. References 1. Beldman, A.; Junfei, B.; Binbin, C.;Zhijun, C.;Xiangming, F .; Wen, D.; Huiyuan, G.; Pei, G.; Beizhong, H.; Dinghuan, H.; et al.White Paper. China Dairy, 1 January 2014; p. 1. 2Gooch, E.; Hoskin, R.; Law, J. China's Dairy Supply and Demand. Livestock, Dairy, Poult . Outlook 2017, 282,27. [CrossRef ]3Gluck, C.; Rentschler, E.; Krewinkel, M.; Merz, M.; von Neubeck, M.; Wenning, M.; Scherer, S.; Stoeckel , M.; Hinrichs, J.;Stressler, T .; et al. Thermostabi l ity of peptidases secreted by microorganisms associated with raw milk. Int. Dairy J. 2016, 56,186-197.I CrossRe f 4. Machado, S.G.; Bagliniere, F.;Marchand, S.; Van Coillie, E.; Vanetti, M.C.D.; De Block, J .; Heyndrickx, M. The Biodiversity of the Microbiota Producing Heat-Resistant Enzymes Responsible for Spoilage in Processed Bovine Milk and Dairy Products. Front.Microbiol. 2017,8,302. [CrossRef ] [PubMed ] 5. Martin, N.; Torres-Frenzel, P.; Wiedmann, M. Invited review: Controlling dairy product spoilage to reduce food loss and waste.J. Dairy Sci. 2021,104,1251-1261. [CrossRef ][PubMed ] 6. Fox, P.F;Uniacke-Lowe, T;McSweeney, P.L.H.;O'Mahony, J.A. Enzymology of Milk and Milk Produc t s; Springer: Berlin/Heidelberg,Germany, 2015; ISBN 9783319148915. 8. Woods, R.G.; Burger, M.; Beven, C.-A.; Beacham, I.R. The aprX-l ipA operon of Pseudomonas fluorescens B52: A molecular analysis of metalloprotease and l ipase production The GenBank accession numbers for the sequences reported in this paper are AF216700, AF216701 and AF216702. Microbiology 2001,147,345-354.[CrossRef ] [PubMed ] 9. Maier, C.; Hofmann, K.; Huptas, C.; Scherer, S.; Wenning, M.; Lucking, G. Simultaneous quantification of the most common and proteolytic Pseudomonas species in raw milk by multiplex qPCR. Appl. Microbiol. Biotechnol. 2021,105,1693-1708. [CrossRef] 10. Zhang, C.; Bijl , E.; Svensson, B.; Hettinga, K. The Extracellular Protease AprX f rom Pseudomonas and its Spoilage Potential for UHT Milk: A Review. Compr. Rev. Food Sci. Food Saf. 2019, 18,834-852. [CrossRef ] 11. Kokkinidou, S.; Peterson, D.G. Control of Maillard-T ype Of f -Flavor Development in Ultrahigh-Temperature-Processed Bovine Milk by Phenolic Chemistry. J. Agric. Food Chem. 2014, 62,8023-8033.[CrossRef ] 12. Dufour, D.;Nicodeme, M.; Perrin, C.; Driou, A.; Brusseaux, E.; Humbert, G.; Gaillard, J.-L.; Dary, A. Molecular typing of industrial strains of Pseudomonas spp. isolated from milk and genetical and biochemical characterization of an extracellular protease produced by one of them. Int . J . Food Microbiol. 2008,125,188-196. [CrossRef ] 13. Datta, N.; Deeth, H. Age Gelation of UHT Milk-A Review. Food Bioprod. Process . Trans . Inst . Chem. Eng. Part C 2001, 79,197-210.I CrossRe f 15. Datta,N.;Kelly, A. The Role of Proteases i n the Stability of UHT-Treated Mi l k. In Agents of Change; Springer: Cham, Switzerland,2021; pp.311-347. ISBN 9783030554828. 16. Mateos, A.;Guyard-Nicodeme, M.; Bagl i niere, F; Jardin, J.;Gaucheron, F.; Dary, A.; Humbert, G.; Gaillard, J .-L. Proteolysis of milk proteins by AprX, an extracellular protease identified in Pseudomonas LBSA1 isolated from bulk raw milk, and implications for the stability of UHT milk. Int. Dairy J. 2015,49,78-88.[CrossRef ] 17. Zhang, W.; Poojary, M.M.; Rauh, V.M.; Ray, C.A.; Olsen, K.; Lund, M.N. Quant i tation of α-Dicarbonyls and Advanced Glycation Endproducts in Conventional and Lactose-Hydrolyzed Ultrahigh Temperature Milk during 1 Year of Storage. J . Agric. Food Chem.2019,67,12863-12874. [CrossRef ] 18. Bagliniere, F.; Tanguy, G.; Jardin, J .; Mateos, A.; Briard, V.; Rousseau, F.; Robert, B.; Beaucher, E.; Humbert, G.; Dary, A.; et al.Quantitative and qualitative variabi l ity of the caseinolytic potential of di f ferent strains of Pseudomonas fluorescens: Implications for the stabi lit y of casein micelles of UHT milks during their storage. Food Chem. 2012, 135,2593-2603.[CrossRef ] 19. Baur, C.; Krewinkel, M.; Kutzli , I .; Kranz, B.; von Neubeck,M.; Huptas, C.; Wenning, M.; Scherer, S.; Stoeckel , M.; Hinrichs, J .; et a l. Isolation and characterisation of a heat-resistant peptidase from Pseudomonas panacis withstanding general UHT processes. Int.Dairy J. 2015, 49, 46-55.[CrossRef ] 20. Stoeckel, M.; Lidolt, M.; Achberger, V.; Gluck, C.; Krewinkel, M.; Stressler, T .; von Neubeck, M.; Wenning, M.; Scherer, S.; Fischer,L.; et al. Growth of Pseudomonas weihenstephanensis, Pseudomonas proteolytica and Pseudomonas sp. in raw milk: Impac t of residual heat-stable enzyme act i vity on stability of UHT milk during shelf-life. I nt. Dairy J. 2016,59,20-28. [CrossRef ] 21. Zhang, C.; Bijl , E.; Hettinga, K. Destabi l ization of UHT milk by protease AprX from Pseudomonas fluorescens and plasmin. Food Chem. 2018,263,127-134. [CrossRef ] 22. Maier, C.; Huptas, C .; Von Neubeck, M.; Scherer, S.; Wenning, M.; Lucking, G. Genetic Organization of the aprX-lipA2 Operon Affects the Proteolytic Potential of Pseudomonas Species in Milk. Front. Microbiol . 2020,11,1190. [CrossRef ] Gaur, V.; Schalk,J.; Anema, S.G. Sedimentation in UHT milk. Int. Dairy J. 2018, 78,92-102. [CrossRef]24 Akkerman, M.;Johansen, L.B.; Rauh, V.; Poulsen, N.A.; Larsen, L.B. Contribution of casein micelle size and proteolysis on protein distribution and sediment formation in UHT milk during storage. Int. Dairy J. 2021, 117, 104980. [CrossRef ] 25. Jansson, T .; Clausen, M.R.; Sundekilde, U.K.; Eggers,N.;Nyegaard, S.; Larsen, L.B.;Ray, C.;Sundgren, A.; Andersen, H.J .; Bertram,H.C. Lactose-Hydrolyzed Milk Is More Prone to Chemical Changes during Storage than Conventional Ultra-High-Temperature (UHT) Mi l k. J. Agric. Food Chem. 2014, 62,7886-7896.[CrossRef] [PubMed ] 26. Nielsen, S.D.; Jakobsen, L.M.; Geiker, N.R.; Bertram, H.C. Chemical l y acidified, live and heat-inactivated fermented dairy yoghurt show distinct bioactive peptides, f ree amino acids and small compounds profiles. Food Chem. 2022,376,131919.[CrossRef ] 29. Pinto, U.M.; Costa,E.D.; Mantovani, H.C.; Vanett i , M. The proteolytic activity of Pseudomonas f luorescens 07A isolated from milk is not regulated by quorum sensing signals. Braz . J . Microbiol.2010,41,91-96. [CrossRef ][PubMed ] 30. Matsel i s, E.; Roussis, I . Proteinase and lipase production by Pseudomonas fluorescens. Proteolysis and l ipolysis in thermized ewe's milk. Food Control 1998,9,251-259. [CrossRef ] 31. Martins, M.; Pinto, U.; Riedel , K.; Vanetti, M.C.D. Milk-deteriorating exoenzymes from Pseudomonas fluorescens 041 i solated from refrigerated raw milk. Braz. J . Microbiol. 2015, 46,207-217.[CrossRef ] 32. Maunsell, B.; Adams, C.;O'Gara,F. Complex regulation of AprA metalloprotease i n Pseudomonas f luorescens M114: Evidence for the involvement of iron, the ECF sigma factor, PbrA and pseudobactin M114 siderophore. Microbiology 2006, 152,29-42.[CrossRe f 33. Anema, S.G. Age Gelation, Sedimentation, and Creaming in UHT Milk: A Review. Compr. Rev. Food Sci. Food Saf. 2018, 18,140-166.I CrossRe f 34. Al-Saadi,J .M.S.; Deeth, H.C. Cross-Linking of Proteins and Other Changes i n UHT Milk during Storage at Different Temperatures.Aust. J. Dairy Technol . 2008,63,93-99. 35. Ranvir, S.; Sharma, R.; Gandhi , K.; Nikam, P;Mann, B. Physico-chemical changes during processing and storage of UHT milk.Indian J . Dairy Sci. 2021, 74,39-47. [CrossRef ] 36. Akkerman, M.; Johansen, L.B.;Rauh, V.; Sorensen, J .; Larsen, L .B.; Poulsen, N.A. Relationship between casein micelle size, protein composi t ion and stability of UHT milk. Int. Dairy J. 2021, 112,104856.[CrossRef ] 37. Li, S.; Ye, A.; Singh, H. Physicochemical changes and age gelation in stored UHT milk: Seasonal variations. Int. Dairy J. 2021, 118, 105028.[CrossRe f 38. Pestana, J.M.; Gennari, A.; Monteiro, B.W.; Lehn, D.N.; Souza, C.F.V. Effects of Pasteurization and Ultra-High Temperature Processes on Proximate Composition and Fatty Acid Prof i le in Bovine Milk. Am. J . Food Technol. 2015,10,265-272. [CrossRef ] 39. Bagliniere, F; Mateos, A.; Tanguy, G.; Jardin,J.; Briard-Bion, V.; Rousseau, F ; Robert, B.; Beaucher, E.; Gaillard, J.L.; Amiel, C.; et a l.Proteolysis of ultra high temperature-treated casein micelles by AprX enzyme from Pseudomonas fluorescens F induces their destabilisation. Int. Dairy J. 2013,31,55-61.[CrossRef ] 40. Stuknyte, M.; Dec i mo, M.; Colzani, M.; Silvetti, T .; Brasca, M.; Cattaneo,S.; Aldini, G.; De Noni, I . Extracellular thermostable proteolytic activity of the milk spoilage bacterium Pseudomonas fluorescens PS19 on bovine caseins. J. Dairy Sc i . 2016, 99, 4188-4195. [CrossRef ] 41. Recio,I.;Garcia-Risco, M.R.; Ramos, M.; Lopez-Fandino, R. Characterization of peptides produced by the action of psychrotrophic proteinases on K-casein. J. Dairy Res. 2000, 67,625-630. [CrossRef ] 42. Reid, J.R.; Coolbear, T.; Ayers, J.S.; Coolbear, K.P. The action of chymosin on k-casein and its macropeptide: Effect of pH and analysis of products of secondary hydrolysis. Int. Dairy J. 1997, 7, 559-569. [CrossRef ] 43. Ramos, M.; Garcia-Risco, M.R.; López-Fandino, R. Proteolysis, protein distribution and stability of UHT milk during storage at room temperature. J. Sc i. Food Agric. 1999,79, 1171-1178. [CrossRef 45. Frohbieter, K.; Ismai l , B.; Nielsen, S.; Hayes, K. Effects of Pseudomonas fluorescens M3/6 Bacterial Protease on Plasmin System and Plasminogen Ac t ivation. J. Dairy Sci. 2005,88, 3392-3401.[CrossRef ][PubMed ] Disclaimer/Publisher's Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or t he editor(s) disclaim responsibility for any injury to people or property resul t ing from any ideas, methods, instructions or products referred to in the content.
确定











还剩13页未读,是否继续阅读?
中国格哈特为您提供《超高温灭菌(UHT)牛奶中蛋白质含量的快速准确检测》,该方案主要用于液体乳中营养成分检测,参考标准《GB 5009.5 食品安全国家标准 食品中蛋白质的测定》,《超高温灭菌(UHT)牛奶中蛋白质含量的快速准确检测》用到的仪器有格哈特杜马斯定氮仪DT N Pro
推荐专场
相关方案
更多
该厂商其他方案
更多
















