方案详情
文
Various techniques yield either structural or functional information about a protein, but DSC uniquely provides insights into both the three-dimensional arrangement and the functional properties of a protein’s structural components.
方案详情

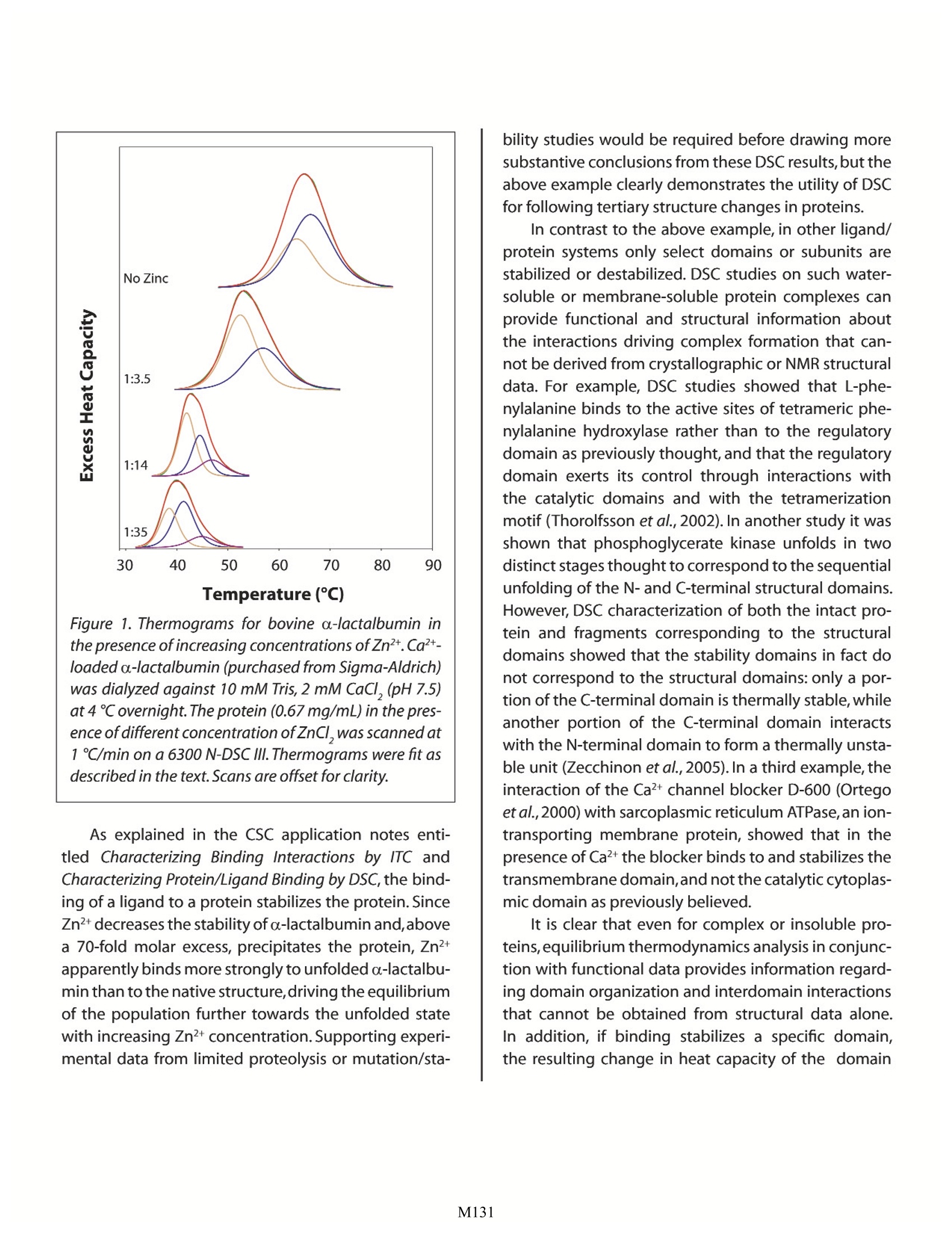
Characterizing Protein Structure by DSC Christin T. Choma TA Instruments, 109 Lukens Drive, New Castle, DE 19720,USA DSC is a valuable approach for characterizing the domain and quaternary structures of proteins. Various techniques yield either structural or functional information about a protein, but DSCuniquely provides insights into both the three-dimensional arrangement and the functionalproperties of a protein's structural components. he non-covalent interactions dictating the threedimensional structures of proteins can be segre-gated into two major categories: those resultingfrom interactions between component amino acids inthe sequence, and those resulting from interactionsbetween the protein and the solvent. These interac-tions, which also control the stability of the protein,aregoverned by thermodynamics.Controlled temperaturechanges have been used extensively to perturb proteinstructures in an effort to learn how heat affects theseinter-andintramolecularinteractions,but it is only in thepast 10 years or so that calorimeters with the sensitivityand baseline stability required for studying very diluteprotein solutions have become commercially available.This note examines the application of differential scan-ning calorimetry (DSC) for monitoring tertiary and qua-ternary structure changes in solubilized proteins, andforms part of a series of application notes dealing withthe biophysical characterization of proteins by DSC. Inparticular, the Calorimetry Sciences Corporation (CSC)application note entitled Characterizing Protein Stabilityby DSC provides a concise description of the thermo-dynamic principles underlying protein stability,and theapplication note entitled Characterizing Protein/LigandBinding by DSC describes the interactions controllingprotein/ligand interactions and their effects on pro-tein stability.For a general description of the principlesbehind DSC, how an experiment is conducted andinterpreted,and a summary of the types of questionsthat can be addressed by this technique, please refer toCSC's Overview Note entitled Life science applications ofDSC. Thermally-induced Structural Changes When globular proteins unfold, significant heat isabsorbed over a temperature range that is characteris-tic for that protein in those specific solvent conditions(pH, protein concentration, buffer salt concentration,etc.). As the temperature of a protein solution is slowlyraised (but prior to the protein unfolding), the baselineobserved on the thermogram increases slightly,then asheating continues, the protein thermally unfolds and gives rise to an endothermic peak. During the unfold-ing process,water molecules around the protein reor-ganize and restructure as hydrophobic residues frominside the protein are exposed; this rearrangement ofwater molecules, coupled with their increasing disor-der as the temperature continues to rise,produces theanomalously large heat capacities of protein aqueoussolutions.Once unfolding is complete,heat absorptiondecreases and a new, higher baseline is established.The contribution of the protein to the calorimetricallymeasured heat capacity (its partial heat capacity) isdetermined by subtracting a scan of a buffer blankfrom the sample data. The data can then be analyzedto provide a complete thermodynamic character-ization of the unfolding process. The area under theendotherm represents the calorimetric enthalpy ofthe unfolding process, the shift in the baseline beforeand after the transition represents the change in heatcapacity of the protein caused by unfolding, and thesharpness of the transition peak is indicative of thecooperative nature of the unfolding process.A narrow,relatively symmetric peak indicates that the transi-tion is probably reversible and highly cooperative.Forsmall single-domain globular proteins,the endothermcan often be accurately represented by a two-statetransition, indicating that the protein unfolds withoutpopulating any intermediate states. The appropriate-ness of the two-state model for a given protein canbe verified by fitting the curve to the two-state van'tHoff equation; this calculation is performed by thesoftware supplied with CSC DSCs. If the van’t Hoff andcalorimetric enthalpies are the same,the denaturationprocess may be accurately approximated by the two-state model. If the van't Hoff enthalpy is smaller thanthe calorimetric enthalpy,an unfolding intermediateis likely formed, whereas if it is larger, the protein likelyassociates and forms oligomers (Marky and Breslauer,1987). The thermodynamic analysis of single domainproteins is described in detail in the CSC applicationnote Characterizing Protein Stability by DSC. A narrow, symmetric endotherm is not neces-sarily indicative of a single-domain protein: a multi- domain protein can give rise to an endotherm thatmimics a two-state transition if all the domains havevery similar thermal stabilities and thus unfold essen-tially simultaneously (Privalov and Privalov,2000). Morecommonly, however, multi-domain proteins produceendotherms that are broad and have a complex shapedue to the independent unfolding of the constituentdomains,each over its own characteristic temperaturerange (Privalov and Potekhin, 1986; Luque et al., 2002).Therefore, the shape of the endotherm encodes ther-modynamic information on the states populated bythe protein throughout the unfolding process. Afterblank subtraction, deconvolution analysis of the endo-therm shows that thermal unfolding proceeds throughseveral cooperative stages, with each stage generally(but not always) corresponding to the unfolding of anindependent structural domain or subunit (Freire, 1995;Privalov and Dragan, 2006). Although tertiary and qua-ternary structure can be inferred from other types ofexperimental information (e.g., homology modeling,limited proteolysis or sequence data), calorimetric dataalone provides a direct measure of the thermodynamicparameters specifying the stabilities of each structuralunit, in addition to information on all the states popu-lated during the unfolding process. Ligand-induced Structural Changes DSC can also be used to characterize both the spe-cific binding of a ligand (for example, a drug to a recep-tor binding site), or nonspecific binding (for example,detergents binding to hydrophobic patches on a pro-tein surface). In some instances ligand binding, evenif to a specific receptor site, results in long-range pro-tein structural rearrangements that stabilize the entirecomplex. For example, the binding of Zn+ to specificsites on insulin triggers the monomeric protein (whichunfolds at 68℃) to oligomerize to a hexameric statethat unfolds at 87C (Huus et al.,2005). DSC can alsoprovide a framework for understanding the thermody-namic basis driving preferential ligand binding: rubre-doxin has been shown to bind metals in the order Fe2+<< Fe3+
确定





还剩4页未读,是否继续阅读?
TA仪器为您提供《蛋白质中蛋白质结构检测方案(差示扫描量热)》,该方案主要用于其他中蛋白质结构检测,参考标准--,《蛋白质中蛋白质结构检测方案(差示扫描量热)》用到的仪器有NANO等温差示扫描量热仪
推荐专场
相关方案
更多
该厂商其他方案
更多
















