中国农业科学院草原研究所 宁夏大学动物科学学院 植物乳杆菌YQM48接种对盐碱土壤中苜蓿青贮饲料品质和微生物群落结构的影响The Effect of Lactobacillus planturum YQM48 Inoculation on the Quality and Microbial Community Structure of Alfalfa Silage Cultured in Saline-Alkali Soil
方案详情

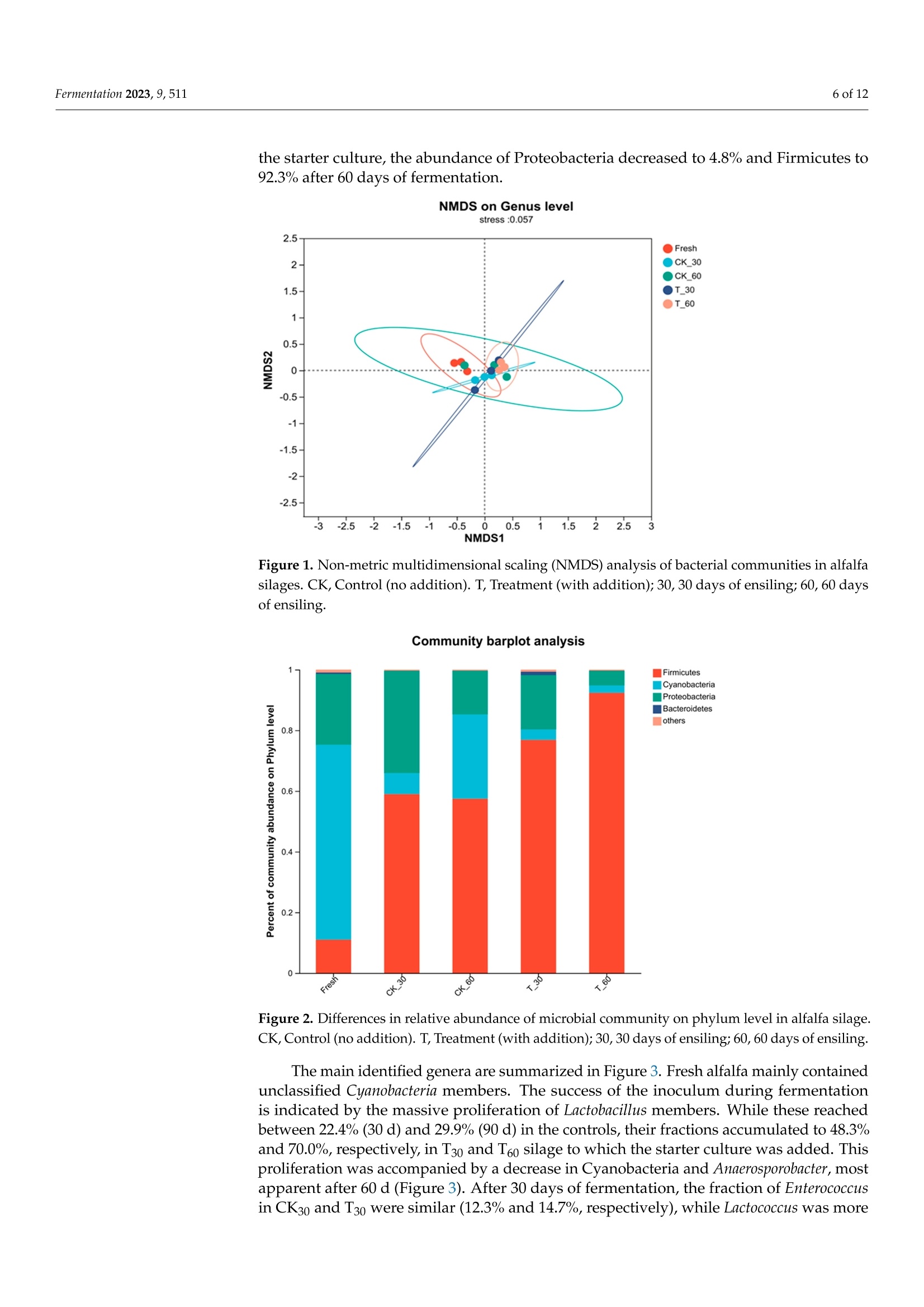
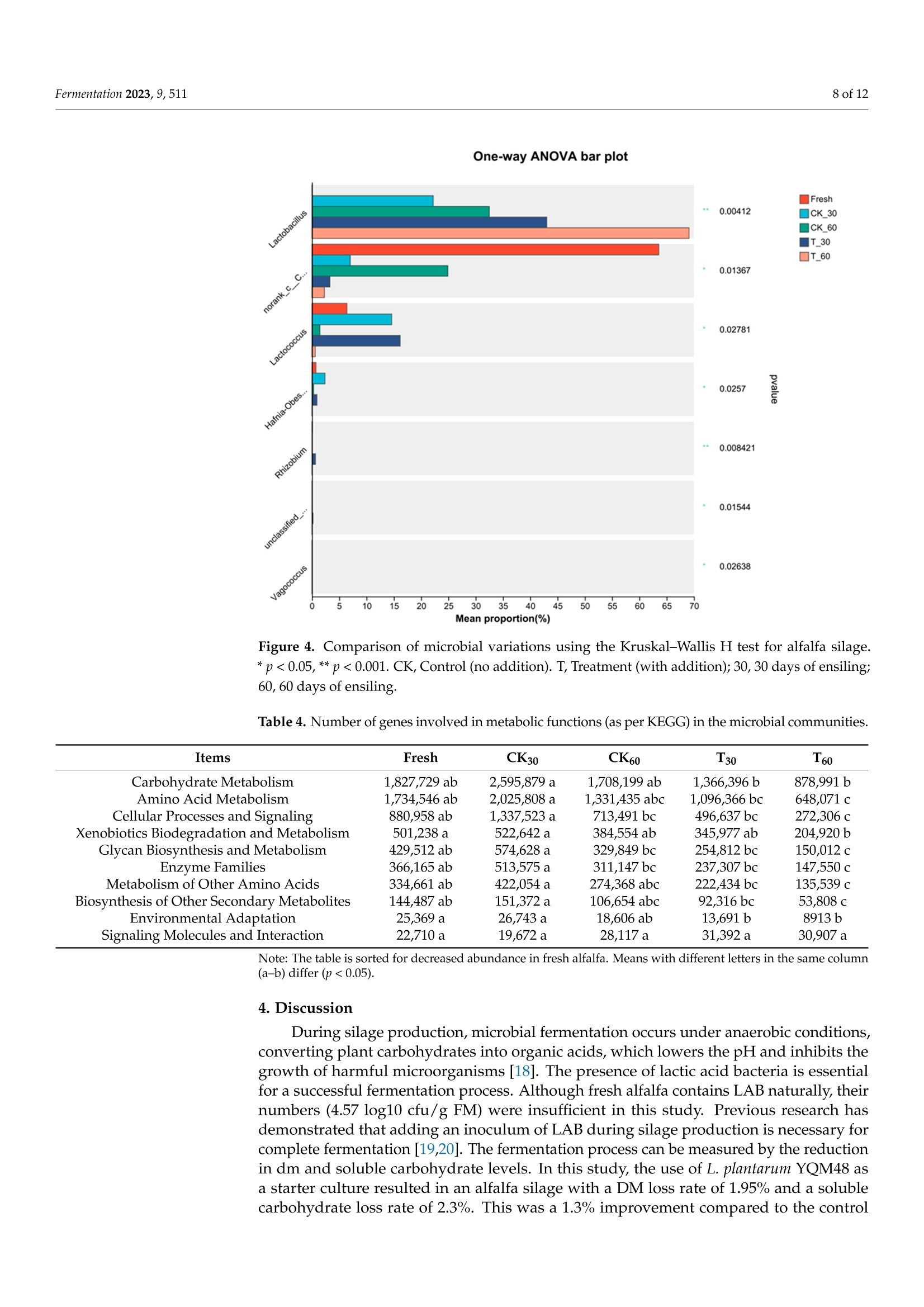
中国农业科学院草原研究所 宁夏大学动物科学学院 植物乳杆菌YQM48接种对盐碱土壤中苜蓿青贮饲料品质和微生物群落结构的影响The Effect of Lactobacillus planturum YQM48 Inoculation on the Quality and Microbial Community Structure of Alfalfa Silage Cultured in Saline-Alkali SoilCharacterization of the Alfalfa and the SilageThe dry matter (DM) of the fresh alfalfa was determined by weight following drying at 65 ◦C for 72 h. Contents of neutral detergent fiber (NDF) and acid detergent fiber (ADF) were measured as per the literature. Water-soluble carbohydrate (WSC) content was determined by colorimetry . The crude protein (CP) was determined using a Kjeldahl (Gerhart Vapodest 50 s, Vapodest, Berlin, Germany) according to AOAC。fermentation 2 of 12Fermentation 2023,9,511 植物乳杆菌YQM48接种对盐碱土壤中苜蓿青贮饲料品质和微生物群落结构的影响 Article The Effect of Lactobacillus planturum YQM48 Inoculation on the Quality and Microbial Community Structure of Alfalfa Silage Cultured in Saline-Alkali Soil Yinghao Liu 1, Yongjie Wang 2, Lianyi Zhang 2, Ling Liu, Ting Cai 2, Chun Chang 1, Duowen Sa1, Qiang Yin1,Xiaowei Jiang 1, Yuyu Li l and Qiang Lu 4* 中国农业科学院草原研究所 1 Inst i tute of Grassland Research, Chinese Academy of Agricultural Sciences, Hohhot 010000, China;liuyinghao@caas.cn (Y.L.); changchun@caas.cn (C.C.); saduowen@caas.cn (D.S.); yinqiang@caas.cn (Q.Y.);jiangxiaowei@caas.cn (X.J.);l i yuyu@caas.cn (Y.L .) 内蒙古农牧推广中心 巴彦淖尔市农业科学研究所 2 Inner Mongolia Agriculture and Animal Husbandry Extension Center, Hohhot 010000, China; 3 wangyongjie@163.com (Y.W.); zhanglianyi312@163.com(L.Z.); caiting@163.com (T.C.) 宁夏大学动物科学学院 Liu, L.; Cai, T.; Chang, C.;Sa,D.; Yin,Q; Jiang, X.; Li, Y.; et al. The Effect of Lactobac il lus planturum YQM48 Inoculation on the Qual i ty and Microbia l Community Structure of Alfalfa Silage Cultured in Saline-Alkal i Soil . Fermentation 2023. 9,511. https://doi .org/10.3390/ fermentation9060511 Academic Editor: Fuyu Yang Received: 4 May 2023 Revised: 24 May 2023 Accepted: 24 May 2023 Published: 26 May 2023 Copyright: @ 2023 by the authors.L i censee MDPI, Basel, Switzerland.This article is an open access article distr i buted under the terms and conditions of the Creative Commons Attribution (CC BY ) l icense (https://creativecommons.org/licenses/by/4.0/) Bayannur City Academy of Agricultura l Sciences, Bayannur 015000, China ; yiyiererwuwu@sina.com 4 Department of Anima l Sc i ence, Ningxia University , Yinchuan 750000, China 米 Correspondence: luqiang@nxu.edu.cn Abstract: Alfalfa cultivated in salt-alkali soil was used for fermentation, to which an inoculum of Lactobacillus plantarum YQM48 was added, to assess its effect on the feed quality and the microbial community structure of the fermented silage. A control was included without inoculum. The nut r itional components, f ermentation quality, pH, and microbial community of the silage were measured after 30 and 60 days of anaerobic fermentation. The results showed that after 30 and 60 days of fermentation in the presence of the inoculum, the content of water-soluble carbohydrates,crude protein content, and dry matter were al l signi fi cant ly higher than those of the control si l age,the pH and butyric acid content were lower, and the content of lactic acid and acetic acid were higher (p<0.05 for all ). There was no significant difference in pH and butyric acid content between 30and 60 days of fermentation in the presence of the inoculum (p > 0.05), while t he lac t ic acid and acetic acid contents were signi f icantly lower in the 60-day silage compared to the 30-day silage (p<0.05). Fermentation reduced the abundance of Cyanobacteria and Proteobacteria (the abundant phyla in the fresh alfalfa), while the abundance of Firmicutes increased, reaching 92.3% after 60days of fermentation with inoculum. The dominant genus in that sample was Lactobac i llus (70.0%),followed by Enterococcus (12.9%), while fermentation of 60 days without i noculum resulted in only 29.2% Lactobacillus, together with 27.8% Cyanobacteria, and 12.2% Enterococcus. In summary, the addition of L . plantarum YQM48 can i mprove the nutritional components and fermentation quality of alfalfa silage cultivated on a salt-alkali soil. Keywords: Lactobacillus plantarum; saline-alkali land; alfalfa; silage quality 1. Introduction Soil where excess salt has accumulated is one of the main types of degraded agricul-tural soils [1]. This type of soil, known as saline-alkali land, is prevalent in 100 countries across six continents and covers a total global area of 960 million hectares, accounting for 9.4% of the total land area, according to UNESCO and FAO [2]. The global area of saline-alkali land i s increasing at a rate of 15 mil l ion hectares per year, resulting in a more than 20% decrease in crop yields annually [3]. The stress and toxicity caused by soil salinity and specific ions severely affect plant growth and development and limit the nutrient accumulation of most agricultural crops [4]. Severe soil salinization can lead to significant losses in crop yields or even total crop failure, hindering the sustainable development of agriculture and posing a major chal l enge for global land management [5]. Therefore, to effectively alleviate t he shortage of arable land, it is necessary to develop and utilize saline-alkali land resources. In the salinization land, alfalfa (Medicago sativa L.) is a valuable source of forage and high-quality protein feed [6]. However, excessive levels of K+, Nat , and Cl - can cause nutrient i mbalances and ionic toxicity. Some plants respond to salt stress by increasing their proline content, while others, such as tomato, increase t heir soluble sugar and protein content [7]. Alfalfa i s i n demand throughout the year and is typically conserved as silage,where the plants undergo natural fermentation [8]. The quality of silage largely depends on lactic acid-producing bacteria (LAB) that create an acidic environment and inhibit the growth of spoilage bacteria [9]. It is crucial to minimize dry matter loss during preservation,and utilizing microbial resources for fermentation is essential to preserve nutrients with minimal energy consumption. Whether the contribution of LAB in silage fermentation can be enhanced is not well explored; for a start, indigenous strains need to be identified that can confer competitive advantages [10]. Culture-independent techniques have already revealed a wide diversity of partially unidentified bacterial species in fermented silage [11]. Whether LAB have the potential to improve silage quality of alfalfa grown under salt stress remains to be estab-lished. In this study, Lactobacillus plantarum strain YQM48 was added as a starter culture to ferment salt-stressed alfal f a, and the s i lage fermentation characteristics and microbial community were studied, to provide a theoretical basis for controlling silage fermentation. 2. Materials and Methods 2.1. Site Description and Alfalfa Production The experimental station where t he alfalfa was grown is located in Ordos City in Inner Mongolia, China, an area in the Hetao Plain (110°37" to 110°27" E and 40°05" to 40°17"N)with a high salinity where an arid, temperate continental climate with strong northwesterly winds dominate. The average annual temperature is 6.8 ℃ with a frost-free period of 165 d,an average annual rainfall of 330 mm, and an average annual evaporation of 2094 mm. The variety of alfalfa tested was the ZhongMu No.3, provided by the Beijing Institute o f Animal Science and Veterinary Medicine (Chinese Academy of Agricultural Sciences,Beijing, China). According to previous studies [6], t his variety i s known for its strong salt resistance, good palatabil i ty, and high nutritiona l value. The plants were sown in May 2020using a drill with a row-to-row distance of 10 cm. 2.2. LAB Strains L. plantarum YQM48 was isolated from plant samples collected from the Salt and Alkaline Land in the Yellow River Basin, Inner Mongolia Autonomous Region, China,which contained 8.5%(w/u) salt. The strains of LAB preserved in our laboratory with good bacteriostasis to Listeria and Escherichia coli were used as the test strains. The name of the strain was YQM48 (NCBI Strain No.OQ592789). The strains to be tested were preserved in nutrient broth liquid medium at -80 °C. The strains were removed from storage and cultured on de Man, Rogosa, and Sharpe (MRS) solid medium at 30°C for 48 h. Then, they were activated for two generations and used in subsequent experiments. After activation,the tested strains were mixed with sterile water to prepare a bacteria l suspension with an OD600 of 0.8, which corresponds to 10° CFU/mL for Lactobacillus grown i n MRS medium. 2.3. Silage Preparation The alfalfa was harvested and left to wilt for 5 h to achieve the desired dry matter content before being cut i nto 2-3 cm f ragments. A specialized fodder chopper was used to chop the resulting material into small, consistent pieces to prevent cross-contamination.To maintain sample integrity, each material was treated separately. The chopped residues were mixed with and without L. plantarum YQM48, which was applied at a rate of 1×10 cfu g -1 of f resh weight in this group. Approximately 2 kg of each replicate was packed into polyethylene plastic bags and vacuum sealed. After 30 and 60 days of ensiling, triplicate samples for each treatment were opened for analysis. 2.4. Characterization of the Alfalfa and the Silage The dry matter (DM) of the fresh alfalfa was determined by weight following drying at 65°C for 72 h. Contents of neutral detergent fiber (NDF) and acid detergent fiber (ADF)were measured as per the l iterature [12]. Water-soluble carbohydrate (WSC) content was determined by colorimetry [13]. The crude protein (CP) was determined using a Kjeldahl (Gerhart Vapodest 50 s, Vapodest, Berlin, Germany) according to AOAC [14]. Following fermentation, 10 g of the produced silage was mixed with 90 mL wa-ter and kept at 4°C for 24 h, after which the l i quid was passed through four layers of cheesecloth and f iltered paper. The f il t rates were used to measure the pH and for deter-mination of ammonium nitrogen [15] and organic acids. The latter were determined by high-performance liquid chromatography (HPLC, Waters e2695,New York, MA, USA) as described before [16]. 2.5. Culture-Dependent Bacterial Enumeration For microbiological characterization, 10 g of each silage sample was added to 90 mL sterile H2O and shaken at 120 rpm for 2 h. For enumeration of dominant microorganisms,ten-fold serial dilutions were prepared, while the original soaking liquid was stored at -80 ℃ for DNA extraction. The dilutions were plated on MRS agar plates for enumeration of LAB, on Violet Red Bile Glucose Agar plates for Enterobacteriaceae, and on potato dextrose agar (Nissui-seiyaku Ltd., Tokyo, Japan) for detection of yeasts. All plates were cultured for 48 h at 37 °C. Aerobic bacteria were determined on nutrient agar medium (Nissui seiyaku Ltd.,Tokyo, Japan). Colony-forming units (CFU) were calculated and reported on a fresh matter (FM) basis. 2.6. High-Throughput Sequencing of the Bacterial Populations Microbial DNA was extracted from the stored soak solution using the E.Z.N.A.@soil DNA Kit (Omega Bio-Tek, Norcross, GA,USA). A control of fresh alfalfa was included in this analysis. After DNA concentration determination by NanoDrop 2000 (Thermo Scien-tific, Wilmington, DE, USA) and verification of t he DNA quality on 1% agarose gels, the DNA was used for PCR ampli f ication o f the V3-V4 variable region of the bacterial 16S rRNA gene using primer 338F and degenerate primer 806R (5'-ACTCCTACGGGAGGCAGCAG and5'-GGACTACHVGGGTWTCTAAT, respectively). The PCR condi t ions involved 3 min of initial denaturation at 95 ℃, followed by 27 cycles of 30 s at 95 °C, 30 s at 55C, and 45 s at 72 °C, with a final 10 min extension at 72 C. PCR reactions were performed in triplicate with 10 ng of template DNA in 20uL and FastPfu Polymerase. The amplicons were purified from a 2% agarose gel using the AxyPrep DNA Gel Extraction Kit (Axygen Bio-sciences, Union City, CA, USA) and quantified using QuantiFluorTM-ST (Promega, Madison,WI,USA). The amplicons were sequenced on an I llumina platform. Following demultiplexing of the raw fastq files and quality filtering with Trimmomatic, the sequences were cleaned and merged by FLASH using the following criteria: (a) reads with a quality score <20 over a 50 bp sliding window were deleted; (b) primer sequences were allowed with a maximum mismatch of 2 nucleotides; (c) reads containing ambiguous bases were removed; and (d)only sequences with an overlap longer than 10 bp were merged. Based on the cleaned and merged sequences, chimeric sequences were identified by UCHIME and removed, after which operational taxonomic units (OTUs) were called with 97% similarity cutoff using UPARSE. The RDP Classifier algorithm was used for taxonomic identif i cation using the Silva database (SSU123) wi t h a confidence threshold of 70%. 2.7. Statistical Analysis The statistical data were analyzed by the procedure of SAS (version 9.3, SAS Institute Inc., Cary, NC, USA). A principal component analysis was carried out using non-metric multidimensiona l scaling analysis (NMDS) to analyze the differences in bacteria l communi-ties in t he various samples. One-way analysis of variance (ANOVA) was conducted using a genera l l inear model in SPSS (version 19.0, IBM Inc., Armonk, NY, USA) to determine the signi f icant difference among samples. The data about chemical and fermentation character-istics, and the abundance of bacteria species of the silages, were analyzed. Mean values were compared using Tukey's test. The level of statistical significance was set to p <0.05. 3. Results 3.1. Characteristics of Fresh Alfalfa Grown on Saline-Alkaline Soil The alfalfa plants grown under saline-stressed conditions were characterized for nutritional quality. The DM content of the plants prior to silage was 29.9%, with a crude protein content of 22.9%, ADF of 33.9%, and NDF content of 37.4%. The fraction of WSC was 7.27%. Culturable bacteria were determined in the wash water of fresh alfalfa. This identified LAB with 4.57 log10 cfu/g FM, and Enterobacteriaceae with 4.8 log1o cfu/g FM.Over a hundred-fold fewer yeast numbers were present, with 2.4 log10 cfu/gFM. 3.2. Effects of L. Plantarum YQM48 on Chemical Composition of Alfalfa Silage The impact of adding an L. plantarum YQM48 inoculum during fermentation on the nutritional quality of the produced silage is summarized in Table 1. After fermentation in the presence of the inoculum, the DM content of the silage was significantly higher than the control, both after 30 and 60 days of fermentation (p<0.05). Likewise, the CP content of the si l age produced i n the presence of the starter culture was higher than the control (p<0.05). The content of ADF only varied slightly, with the lowest content reported for CK60, and that t reatment also resulted in the lowest NDF content. The difference in NDF content between samples fermented without and with the added bacteria was significant at both time points (p<0.05). Lastly, the content of WSC was significantly higher when fermentation was performed in t he presence of L. plantarum YQM48(p<0.05). Table 1. Chemical composition of alfalfa silage after 30 d and 60 d of ensiling. Items CK30 T30 CK60 T60 Dry matter (DM) (weight %) 27.5±0.24b 28.7±0.12a 27.3±0.12b 28.6±0.22a Crude protein (CP,%DM)* 19.5±0.05b 21.2±0.17a 19.0±0.17c 21.9±0.12a Acid detergent fiber (ADF,% DM)* 31.6±0.12a 31.7±0.09a 31.2±0.21b 31.5±0.05 ab Neutral detergent fiber (NDF,% DM)* 35.7±0.05b 36.3±0.08a 35.2±0.12c 36.3±0.08a Water soluble carbohydrates (WSC,%DM)* 5.03±0.21c 5.15±0.05a 5.01±0.25c 5.07±0.05b CK, Control (no addition). T , Treatment (with addition). * Percentage expressed per dry weight. All data are reported as average wi t h standard variation based on t riplicates. Means with different letters in t he same column (a-c) di f fer (p<0.05). 3.3. Effects of L. plantarum YQM48 on Fermentation Characteristics of Alfalfa Silage The fermentation quality of the produced silage was assessed by pH, organic acid content, and ammonium nitrate content (Table 2). The pH was lower after 60 d of fermenta-tion than after 30 d in absence of the starter culture, and addition of the L. plantarum strain reduced the pH compared to the control , at both time points (p <0.05). HPLC analysis detected lactic acid (LA), acetic acid (AA), and butyric acid (BA), but no propionic acid. The latter is known to negatively affect palatability of silage [17]. The LA content was signifi-cant l y higher in the silage produced with the starter culture t han in t he control, and higher after 60 d than after 30 d fermentation with the inoculum (p <0.05) (Table 2). A similar trend was observed for the AA content of the samples, while the BA content was generally lower than the other two acids and was much lower in t he silage produced with the starter culture than in the control. The amount of ammonium nitrogen was much lower in the silage produced with the starter culture compared to the control, tho O u u gh no differences were observed between 30 or 60 days of fermentation (Table 2). The results showed that the addition of a starter culture of L. plantarum YQM48 improved the fermentation quality of the alfalfa si l age. Table 2. Fermentation characteristics of alfalfa silage after 30 d and 60 d of ensiling. Items CK30 T30 CK60 T60 pH 5.1±0.05a 4.4±0.09c 4.7±0.05b 4.3±0.17c Lactic acid (LA,%DM)* 3.7±0.05c 4.5±0.12b 3.6±0.12c 4.9±0.05a Acetic acid (AA, %DM)* 3.1±0.05b 3.5±0.05b 3.3±0.05c 3.8±0.01a Butyric acid (BA,%DM)* 1.5±0.14ab 1.3±0.18bc 1.7±0.09a 1.2±0.01c NH4+-N(%DM) 3.0±0.13a 1.9±0.12b 3.1±0.05a 1.8±0.1b CK, Control (no addition). T, Treatment (with addition). *Percentage expressed per dry weight . Al l data are reported as average wi t h standard variation based on t riplicates. Means with different letters in the same column (a -c) differ (p<0.05). 3.4. Characterization of the Bacterial Communities in Fresh Alfalfa and in the Silage Following amplification and sequencing of t he V2-V3-variable region of the bacterial 16S rRNA gene, the bacterial microbiome of the fresh and fermented alfalfa was f i rst characterized by diversity index analysis (Table 3). The coverage of all samples was above 99%. The OTU index in the silage varied from 94 (T30, CK30) to 106 (T60), which was lower than the 114 OTUs identi f ied in the fresh alfalfa . This indicates that during fermentation, a group of organisms became dominant that reduced the numbers of others. This can also be seen from the reduced Shannon, Ace, and Chao 1 indices. Both fermentation time and the addition of the starter culture resulted in a reduced Alpha diversity of the bacteria. Table 3. Alpha diversity of the bacterial community in fresh materials and alfalfa silage. Treatment OTU Shannon Ace Chao 1 Coverage Fresh 114.7a 1.6b 125.4a 127.7a 0.99 a T30 94 ab 2.6a 108.6 ab 105.1 ab 0.99 a CK30 94 ab 2.1ab 95.1b 90.8b 0.99 a T60 CK60 106.3 ab 2.2 ab 116.6 ab 115.5 ab 0.99 a 81.3b 1.9 ab 98.8b 99.5 ab 0.99 a CK, Control (no addition). T, Treatment (with addition). Means with di f ferent letters in the same column (a-b)dif f er (p<0.05). The identified bacterial populations were compared by principal component analysis (Figure 1). There was considerable variation between the triplicates of the CK60 samples,and to a lesser extent i n t he T30 samples. As shown in the f igure, the value of the applied NMDS analysis is 0.057, less than 0.1, indicating that the analysis is relatively complete.There was no overlap between CK30 and CK60 treatments, suggesting that the microbial community structure was quite different between these two t ime points. This indicates a highly instable and variable community during fermentation in the absence of a starter culture, which may follow an unorganized and stochastic path. However, there was also no overlap between T30 and T60 treatments, suggesting that even in the presence of the starter culture, the difference in bacterial community structure strongly varied over fermentation time. The composition of the bacterial communities was analyzed at the phylum level, with findings summarized in Figure 2. In fresh alfalfa, members of the phylum Cyanobacteria dominated , with 64.2%, and Proteabacteria accounted for 23.3%, while Firmicutes, to which LAB belong, were present at 9.9% only. Fermentation largely promoted the presence of Firmicutes , and an increase in this fraction from 30 to 60 days was noted in the treatment samples. In the control silage, the abundance of Proteobacteria decreased to 14.4% after 60 days of fermentation while Firmicutes increased to 57.5%. In the silage produced with the starter culture, the abundance of Proteobacteria decreased to 4.8% and Firmicutes to 92.3% after 60 days of fermentation. NMDS on Genus level F re sh C K 30 C K 60 T _30 T _60 Figure 1. Non-metric multidimensional scaling (NMDS) analysis of bacterial communities in alfalfa silages. CK, Control (no addition). T, Treatment (with addition); 30, 30 days of ensi li ng; 60, 60 days of ensiling. Community barplot analysis Figure 2. Differences in relative abundance of microbial community on phylum level in alfalfa silage.CK, Control (no addition). T, T reatment (with addition); 30, 30 days of ensiling; 60, 60 days of ensiling. The main identi f ied genera are summarized i n Figure 3. Fresh alfal f a mainly contained unclassified Cyanobacteria members. The success of the inoculum during fermentation is indicated by the massive proliferation of Lactobacillus members. While these reached between 22.4% (30 d) and 29.9%(90 d) in the controls, their fractions accumulated to 48.3%and 70.0%, respectively, in T30 and T60 silage to which the starter culture was added. This proliferation was accompanied by a decrease in Cyanobacteria and Anaerosporobacter, most apparent after 60 d (Figure 3). After 30 days of fermentation, the fraction of Enterococcus in CK30 and T30 were similar (12.3% and 14.7%, respectively), while Lactococcus was more abundant in CK30 (20.2%) than in T30 (13.7%). That latter genus reduced in numbers after longer fermentation, espec i ally i n the presence of the i noculum, where i t only represented 2.5% in T60. The results indicate that the inoculum had not only proliferated members of the Lactobacillus but had also caused shifts in other dominant genera. Community barplot analysis Figure 3. Bacterial community composition on genus level in alfalfa silage. CK, Control (no addition).T, Treatment (with addi t ion); 30, 30 days of ensiling; 60, 60 days of ensiling. Signi f icant shif t s i n the abundance of bacterial genera are summarized i n Figure 4. The strongest shifts were observed for Lactobacillus, as expected, but the shifts in Lactococcus were also substantial . Other statistically significant shifts i nvolved genera that repre-sented minor fractions only, including Hafnia-Obesumbacterium, Rhizobium, unclassified Xanthomonadaceae, and Vagococcus (Figure 4). The identified genera were used to infer microbial metabolic pathways in which the dominant bac t eria may be involved . For this, gene IDs corresponding to each OTU in the microbiomes were identified and classified into various pathways and functional infor-mation using the KEGG database. The pathway for which most genes were identified was the Carbohydrate Metabolism, though 60 d of fermentation reduced these numbers (Table 4). The second most abundant pathway for which genes were i dentified was the Amino Acid Metabolism, but again, numbers decreased with fermentation (Table 4). Other main microbial functions included cellular Processes and Signaling, Xenobiotics Biodegra-dation and Metabolism, and Glycan Biosynthesis and Metabolism. When the fraction of genes involved in a given pathway was considered, it was apparent that genes involved i n Cellular Processes and Signaling significantly decreased during fermentation compared to fresh alfalfa, with lower fractions after 60 d of fermentation compared to 30 d, and with lower fractions in the treatment samples compared to the controls. T Fre sh J C K 30 C K _60 T _30 T _60 Figure 4. Comparison of microbia l variations using the Kruskal -Wal li s H test for alfalfa silage.* p <0.05, **p<0.001. CK, Control (no addition). T, Treatment (with addition); 30, 30 days of ensiling;60, 60 days of ensiling. Table 4. Number of genes involved in metabolic functions (as per KEGG) in the microbial communities. ltems Fresh CK30 CK60 T30 T60 Carbohydrate Metabolism 1,827,729 ab 2,595,879 a 1,708,199 ab 1,366,396 b 878,991 b Amino Acid Metabolism 1,734,546 ab 2,025,808 a 1,331,435 abc 1,096,366 bc 648,071c Cellular Processes and Signaling 880,958 ab 1,337,523 a 713,491bc 496,637 bc 272,306c Xenobiotics Biodegradation and Metabolism 501,238 a 522,642 a 384,554 ab 345,977 ab 204,920b Glycan Biosynthesis and Metabolism 429,512 ab 574,628 a 329,849 bc 254,812 bc 150,012c Enzyme Families 366,165 ab 513,575 a 311,147 bc 237,307 bc 147,550c Metabolism of Other Amino Acids 334,661 ab 422,054 a 274,368 abc 222,434 bc 135,539c Biosynthesis of Other Secondary Metabolites 144,487 ab 151,372 a 106,654 abc 92,316 bc 53,808c Environmental Adaptation 25,369 a 26,743 a 18,606 ab 13,691b 8913b Signaling Molecules and Interaction 22,710 a 19,672 a 28,117a 31,392 a 30,907 a Note: The table is sorted for decreased abundance in fresh alfalfa. Means with different letters in the same column (a-b) differ (p<0.05). 4. Discussion During silage production, microbial fermentation occurs under anaerobic conditions,conver t ing plant carbohydrates into organic acids, which lowers t he pH and inhibits the growth of harmful microorganisms [18]. The presence of l actic acid bacteria is essential for a successful fermentation process. Although fresh alfalfa contains LAB naturally, their numbers (4.57 log10 cfu/g FM) were insufficient in this study. Previous research has demonstrated that adding an inoculum of LAB during silage production is necessary for complete fermentation [19,20]. The fermentation process can be measured by the reduction in dm and soluble carbohydrate levels. In this study, the use of L. plantarum YQM48 as a starter culture resulted in an alfalfa silage with a DM loss rate of 1.95% and a soluble carbohydrate loss rate of 2.3%. This was a 1.3% improvement compared to the control group's DM reduction. The decrease in DM and carbohydrates is due to the metabolic activity of lactobacilli during fermentation. These microorganisms convert nutrients into organic acids, ethanol, and other metabolic products [21,22]. The use of L. plantarum starters during silage production ensures sufficient lactic acid production in the early stages of fermentation, which rapidly lowers the pH and inhibits the growth of putrefactive microorganisms [31,32]. Our study demonstrated an increase in lactate and acetate production following inoculation, resulting in a lower pH similar to that reported in literature [33]. The slightly higher pH observed i n the CK30 group, which served as the control silage group, is not unexpected. This is because the control group did not receive any treatment or intervention, and therefore, the natural fermentation process may have resulted i n a slightly higher pH compared to the experimental groups. This observation is consistent with previous studies that have reported higher pH values in control silage groups [33,34]. Overall, t he slightly higher pH in the control group does not affect the validity of the study results, as the experimental groups were compared to each other rather than to the control group. After 60 days of fermentation, Lactobacillus was the dominant bacteria, whereas controls without inoculation contained more miscellaneous bacteria such as Cyanobacte-ria. The abundance of L. plantarum can reduce the ammonium nitrogen content in silage feed [34]. Longer fermentation times increase the abundance of lactobacilli, and the acidic environment they create reduces the propagation of miscellaneous and mold microorgan-isms [35], as observed in our study. The reduced Shannon index of microbial community during inoculated fermentation suggests that silage additives can reduce the diversity and richness of bacterial communities [36]. Before silage, the main bacterial phyla were Cyanobacteria, Proteobacteria, and Fir-micutes (in that order). However, after fermentation, Firmicutes vastly outnumbered the other phyla, which has also been observed by others [37]. The dominant bacterial genera during fe rmentation without i noculum changed from Lactobacillus, Enterococcus,Lactococcus,and Enterobacter (in decreasing order) at day 30 to Lactobacillus, Cyanobacteria, Enterococcus,and Anaerosporobacter after 60 days. Pantoea (which did not proliferate in our samples)and Enterobacteriaceae can produce acetic acid, propionic acid, and succinic acid through carbohydrate metabolism under anaerobic conditions, and such organisms may reduce the carbohydrate content [38]. In contrast, Lactobacillus can decompose soluble carbohydrates to form lactate through glycolysis or by the hexose phosphate pathway. Clearly, the high abundance of Lactobacillus enabled by the i noculum increased the l actate content , and after 60 days of fermentation, the soluble carbohydrates had decreased by 2%. This i llustrates how anaerobic fermentation was enhanced as a result of the inoculum. 5. Conclusions The addition of a L. plantarum YQM48 inoculum to alfalfa grown in a saline-alkali soil significantly improved t he quality of the produced silage. After 60 days of fermentation, the CP content, pH value, and lactic acid content of the silage were significantly higher than in silage produced without inoculum. The addition of the strain decreased the bacteria l diver-sity and changed the community structure of f ermented silage, with Lactobacillus becoming highly dominant, while fermentation without the inoculation allowed the proliferation of Cyanobacteria and other genera. Author Contributions: Methodology, Y.W., C.C., Y.L. (Yuyu Li); Validation, Q.Y.; Formal analysis,D.S.; I nvestigation, L.L. and T.C.; Resources, L.Z.; Data curation, X.J.; Writing -original draft, Y.L .(Yinghao Liu) and Q.L. Al l authors have read and agreed to the published version of the manuscript. Funding: This work was supported by t he Ningxia Higher Education Institutions F i rst-Class Disci-pline Construction Project (NXYLXK2017A01), the Fundamental Research Funds of Chinese Academy of Agriculture (110233160007007), Inner Mongolia Autonomous Region science and technology plan-ning project (2022YFHH0046), Inner Mongolia Natural Science Foundation project (2022LHQN3003). Institutional Review Board Statement: Not applicable Informed Consent Statement: Not applicable. Data Availability Statement: The datasets presented in this study can be found in online repositories.We uploaded the sequences data i n the NCBI and got an accession number PRJNA753242. Conflicts of Interest: The authors declare no conflict of interest. References 1. Shao, H.; Chu, L.; Lu, H.; Qi, W.; Chen, X.; Liu, J.; Kuang,S.; Tang, B.; Wong, V. Towards sustainable agriculture for the salt-affected soil. Land Degrad . Dev.2018,30,574-579. [CrossRef ] 2. Li, C.; Wang, Z.; Xu, Y.; Sun, J;Ruan, X.; Mao, X.;Hu, X.; Liu, P. Analysis of the effec t of Modi f ied biochar on saline-alkal i soil remediation and crop growth. Sustainability 2023, 15,5593. [CrossRef ] 3. Zhang, L.;Ge, A.; An, F.;Guo, L.; Nie, Z.; Liu, J; Yang, F; Wang, Z. Soi l bacterial microbiota predetermines rice yield in reclaiming saline-sodic soils l eached with brackish ice. J . Sci. Food Agric. 2021, 101, 6472-6483. [CrossRef ] 4 He, F; Wang, G.; Wang, L.; Li, Z.; Tong, Z.; Wang, Y.; Li, X. Effects of organic base fertilizer and inorganic topdressing on al f al f a productivity and the soi l bacterial community i n saline soil of the huanghe r iver delta in China. Agronomy 2022,12,2811.[CrossRef ] 5. Zhao, Y.; Zhang, Z.; Li, Z.; Yang, B.; Li, B.; Tang, X.; Lai, Y. Comprehensive study on saline-alkali soil amelioration with sediment of irrigation area in northeast China. Arab. J. Chem. 2023, 16,104608. [CrossRef ] 7. Liu, H.; Lu, X.; Li, Z.;Tian, C.; Song, J. The role of root-associated microbes i n growth stimulation of plants under saline conditions.Land Degrad. Dev. 2021, 32,3471-3486. [CrossRef ] 9. Yang, C.; Wang, X.; Miao, F; Li, Z.; Tang, W.; Sun, J. Assessing the effect of soil salinization on soil microbial respiration and diversities under incubation conditions. Appl . Soil Ecol. 2020, 155,103671.[CrossRef] 10. Zhang, H.; Gao, S.; Zhang, X.; Meng, N.; Chai, X.; Wang, Y. Fermentation characteristics and the dynamic trend of chemical components during fermentation of massa medicata dermentata. Arab. J. Chem. 2021, 15, 103472. [CrossRef ] 11. Dong , Z.; Yuan, X.; Wen, A.; Desta, S.T.; Shao, T . Effects of calcium propionate on the fermentation quality and aerobic stability of alfalfa silage. Asian-Australas. J . Anim. Sci. 2017,30,1278-1284. [CrossRef ] [PubMed ] 12. Van, S.;Robertson,J; Lewis, B. Methods for dietary fiber, neutral detergent fiber, and nonstarch polysaccharides in relation to animal nutri t ion. J. Dairy Sci. 1991,74,83-97. 13. Lei , C.; Gang, G.; Cheng, Q.; Jie, Z.;Masataka, S.; Tao, S. The effects of replacement of whole-plant corn with oa t and common vetch on the fermentation quality, chemical composi t ion and aerobic stability of total mixed ration silage in Tibet. Anim. Sci. 2015,86,69-76. 14. Gao, W.; Ynag , G.;Gao, X.; Yu, Z.;Xu, Q.; Yuan, X. Effects of nitrogen, phosphorus and potassium fertilizer on yield and quality of silage maize. Crop J. 2018, 21,144-149. 15. Hao,J; Yu, H.; J ia , Y.; Wang, Z.; Ge, G.; Sun, L. Effects of silage density and silage t ime on fermentation quality and nutrient composition of alfalfa. J. Grassl . Sci. 2022, 30,2492-2496. 16. Si, Q.; Wang, Z.; Liu, W.; Liu, M.; Ge, G.; Jia, Y.; Du, S. Influence of cellulase or Lac t iplantibacillus plantarum on the ensiling performance and bacterial community i n mixed silage of alfalfa and l eymus chinensis. Microorganisms 2023, 11, 426. [CrossRef ] 17. Hao, W.; Tian, P .; Zheng, M.; Wang, H.; Xu, C . Characteristics of proteolytic microorganisms and their effects on proteolysis in total mixed ration silages of soybean curd residue. J. Anim. Sci. 2020, 33,100-110. [CrossRef] [PubMed ] 18. Silva, V.P.;Pereira, O.G.; Leandro,E.S.; Da Silva, T.C.; Ribeiro, K.G.; Mantovani, H.C.; Santos, S.A. Effects of lactic acid bacteria with bacteriocinogenic potential on the fermentation profile and chemical composition of alfalfa silage in tropical conditions.J. Dairy Sci . 2016, 99,1895-1902.[CrossRef ] 19. Hisham, M.B.; Hashim, A.M.; Hanafi,N.M.;Rahman, N.A.; Tan, C.K.; Nazli , M.H. Bacterial communities associated with silage of different forage crops in Malaysian climate analysed using 16S ampl i con metagenomics. Sci. Rep. 2022, 12,7107. [CrossRef ] 20. Huo, W.; Zhang, Y.; Zhang, L.; Shen, C .; Zhang, S.; Wang, C.; Guo, G. Effec t of Lactobacilli inoculation on protein and carbohydrate fractions, ensiling characteristics and bacterial community of alfalfa silage. Front. Microbiol. 2022,13,1070175.[CrossRef ] 21. Fan, X.; Xie, Z.; Cheng, Q.; Li, M.; Long, J; Lei, Y.; Jia, Y.; Wang, Z. Fermentation quality, bacterial community, and predicted functional profiles in silage prepared with alfalfa, perennia ryegrass and their mixture in the karst region. Front. Microbiol . 2022,13,1062515.[CrossRe f 22. Bao, X.; Feng, H.; Guo, G.; Huo, W.; Li, Q.; Xu, Q.; Liu, Q.; Wang, C.; Chen, L. Effects of laccase and lactic acid bac t eria on the fermentation qual i ty, nutrient composition, enzymatic hydrolysis, and bacterial community of alfalfa silage. Front . Microbiol.2022,13,1035942. [CrossRef ] [PubMed ] 23. Si, H.; Liu, H.; Li, Z.; Nan, W.; Jin, C.; Sui, Y.; Li, G. Effect of Lactobacillus plantarum and Lactobacillus buchneri addition on fermentation, bacterial community and aerobic stability i n l ucerne silage. Anim. Prod. Sci . 2018,59,1528-1536. [CrossRef ] 24. Yang, F; Wan, Y.;Zhao, S.; Wang, Y. Lactobac i llus plantarum inoculants delay spoilage of high moisture alfalfa silages by regulating bacterial community composition. Front . Microbiol. 2020, 11, 1989.[CrossRef ] [PubMed ] 25. Guo,J.; Xie, Y.; Yu, Z.;Meng, G.; Wu, Z. Effect of Lactobacillus plantarum expressing multifunctional glycoside hydrolases on the characteristics of alfalfa silage. Appl. Microbiol. Biotechnol. 2019, 103, 7983-7995. [CrossRef ] 26..Chen, Q.; Li , M.; Fan, X.; Chen, Y.; Sun, H.; Xie, Y.; Zheng, Y.; Chen, C.; Li , P. Effects of epiphytic and exogenous lactic acid bacteria on fermentation quality and microbial community compositions of paper mulberry silage. Front. Microbiol. 2022,13,973500. [CrossRef ] 27. Su, R.; Ni , K.; Wang, T.; Yang, X.; Zhang, J.; Liu, Y.; Shi, W.; Yan, L.; Jie, C. Effects of ferulic acid esterase-producing Lactobacillus fermentum and cellulase additives on the fermentation quality and microbial community of alfal f a silage. PeerJ 2019,7,e7712.[CrossRef |[PubMed ] 28. Wang, C .; He, L.;Xing, Y.; Zhou, W.; Yang, F.;Chen, X.; Zhang, Q. Fermentation quality and microbial community of alfalfa and stylo si l age mixed with moringa oleifera leaves. Bioresour. Technol. 2019, 3,129.[CrossRef ] 29. Bai , B.; Qiu , R.; Wang, Z.; Liu, Y.; Bao,J; Sun, L.; Liu, T.; Ge, G.; Jia, Y. Effects of cellulase and l actic acid bacteria on ensiling performance and bacterial community of Caragana korshinskii silage. Microorganisms 2023, 11, 337. [CrossRef ] 30. Fan, X.; Zhao, S.; Yang, F; Wang, Y. Effects of lactic acid bacterial inoculants on fermentation quality, bacterial community, and mycotoxins of alfalfa silage under vacuum or nonvacuum treatment. Microorganisms 2021, 9,2614.[CrossRe f ] 31. He, L.; Wang, Y.; Guo, X.; Chen, X.; Zhang, Q. Evaluating the effectiveness of screened lactic acid bacteria in improving crop residues silage: Fermentation parameter, nitrogen fraction, and bacterial community . Front. Microbiol . 2022,13,680988. [CrossRef ][PubMed l 32. Na, N.;Moge, Q.; Wu, N.; Sun, L.; Xu, H.; Zhao, Y.; Wei, X.; Xue, Y.; Tao, Y. Bacterial community and fermentation qual i ty of ensi l ing alfalfa with commercia l lactic acid bacterial additives. Front . Microbiol. 2022,13,836899. [CrossRef ][PubMed] 33. Silva, V.P.; Odilon, G.P.; Eliana, S.L.; Rosinea, A.P.; Mariele, C.N.; Karina, G.R. Selection of lactic acid bacteria from alfalfa silage and i ts effects as inoculant on silage fermentation. Agriculture 2020, 10,518. [CrossRef ] 34. Xie, Y.; Bao, J.; Li, W.; Sun, Z.; Gao, R.; Wu, Z.; Yu, Z. Effects of applying lactic acid bacteria and molasses on the fermentation quality, protein fract i ons and in vitro digestibility of baled alfalfa silage. Agronomy 2021, 11,91. [CrossRef ] 37. Zhao, S.; Wang, Y; Yang, F; Wang, Y;Zhang, H. Screening a Lactobacillus plantarum strain for good adaption in alfalfa ensiling and demonstrating its improvement of alfalfa silage quality. J. Appl . Microbiol. 2020,129,233-242. [CrossRef ] [PubMed ] 38. Yuan, X.; Li,J;Dong, Z.; Shao, T. The reconstitution mechanism of napier grass microiota during the ensiling of alfalfa and their contributions to fermentation quality of si l age. Bioresour. Technol. 2019, 297,122391. [CrossRef ] Disclaimer/Publisher's Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, method s , i nstructions or products referred to in the content.
确定









还剩10页未读,是否继续阅读?
中国格哈特为您提供《接种植物乳杆菌YQM48的苜蓿青贮饲料粗蛋白、中性洗涤纤维、酸性洗涤纤维含量的检测》,该方案主要用于畜牧中动物营养与健康检测,参考标准--,《接种植物乳杆菌YQM48的苜蓿青贮饲料粗蛋白、中性洗涤纤维、酸性洗涤纤维含量的检测》用到的仪器有格哈特带自动进样器凯氏定氮仪VAP500C、格哈特红外加热消解快速消化系统TTs125、格哈特自动升降凯氏定氮电热消解仪KT-L 20s、格哈特全自动型纤维分析仪FT12、德国加液器MM、格哈特维克松废气实验室废物处理系统涤气VS、凯氏定氮催化片
相关方案
更多
该厂商其他方案
更多