
方案详情
文
德国默克(Merck)的科学家们设计了基于牛CDR-H3双特异性抗体,靶向肿瘤细胞上的表皮生长因子受体(EGFR)以及自然杀伤(NK)细胞上的天然细胞毒性受体NKp30。基于牛IgG的双特异性抗体(BsAbs)引发了有效的NK细胞重定向,从而引发了EGFR过表达的肿瘤细胞裂解以及促炎细胞因子的强劲释放[2]。
方案详情

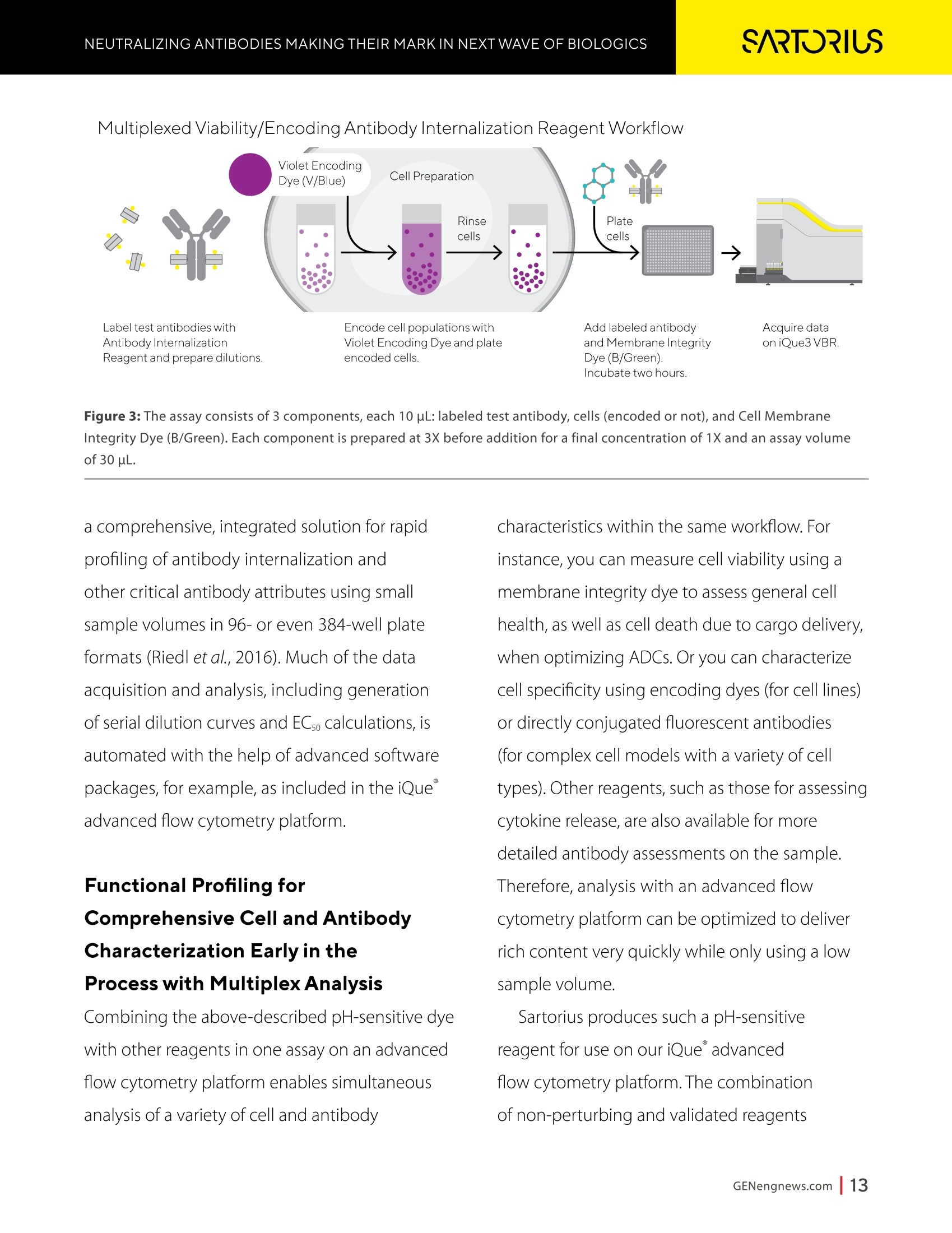
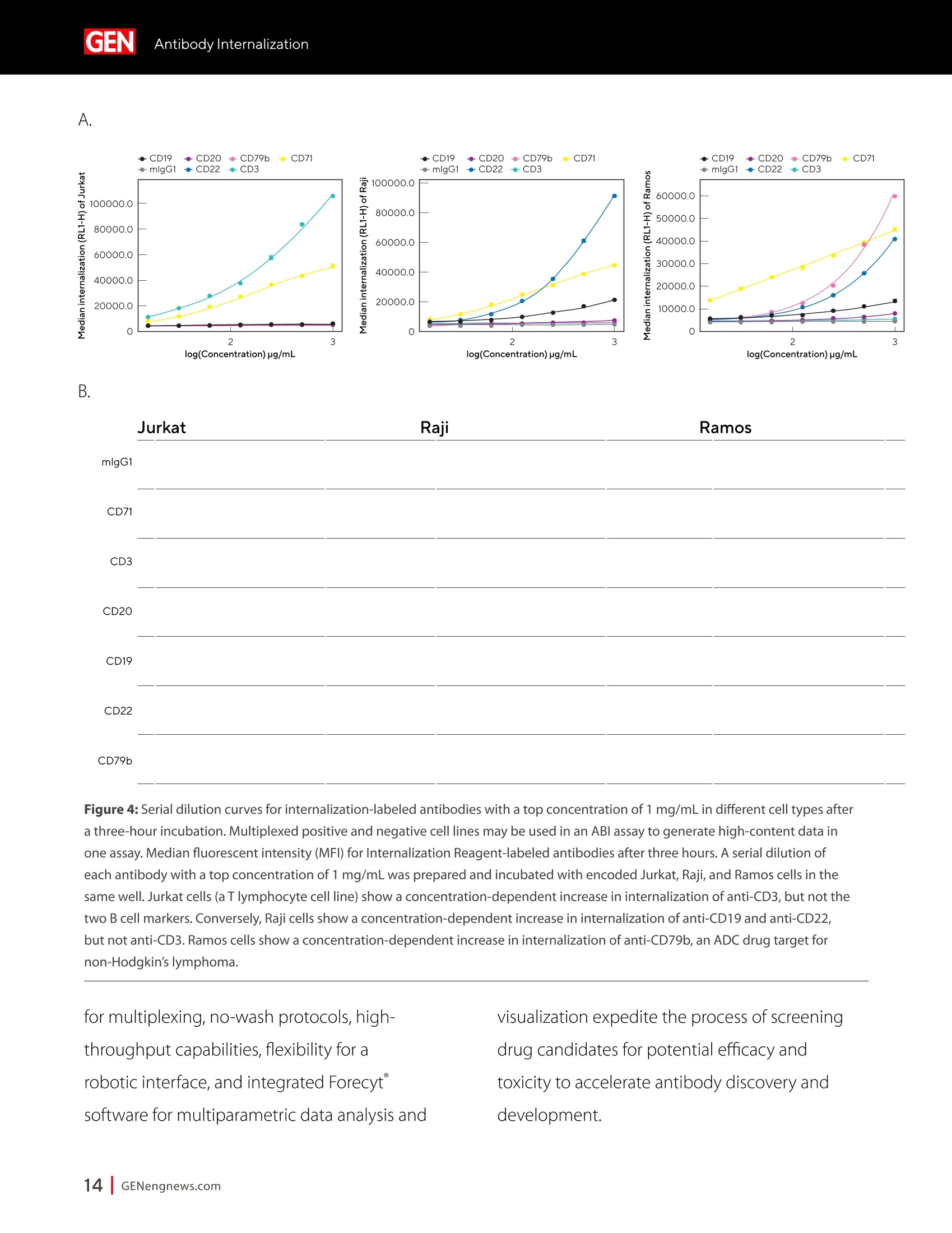
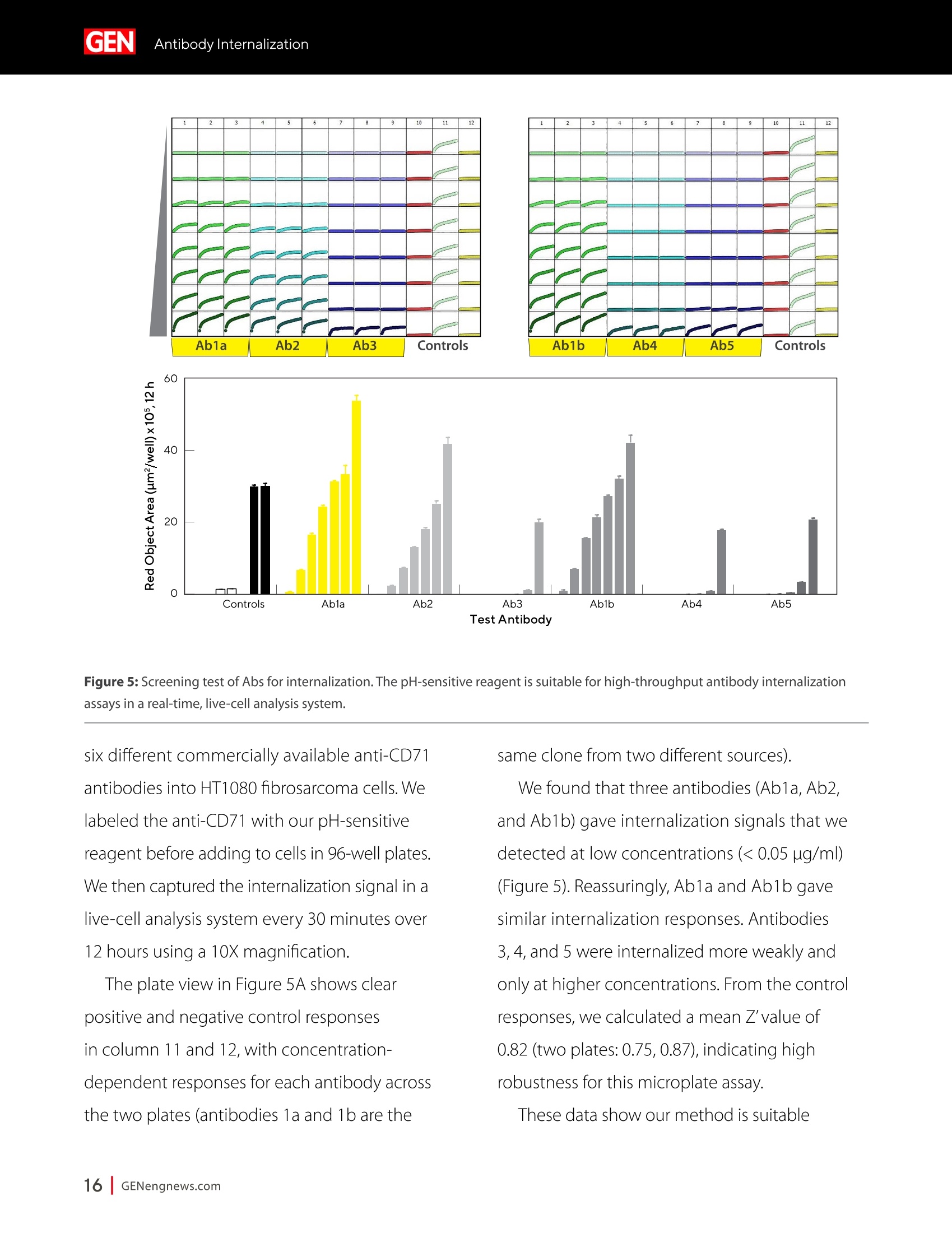
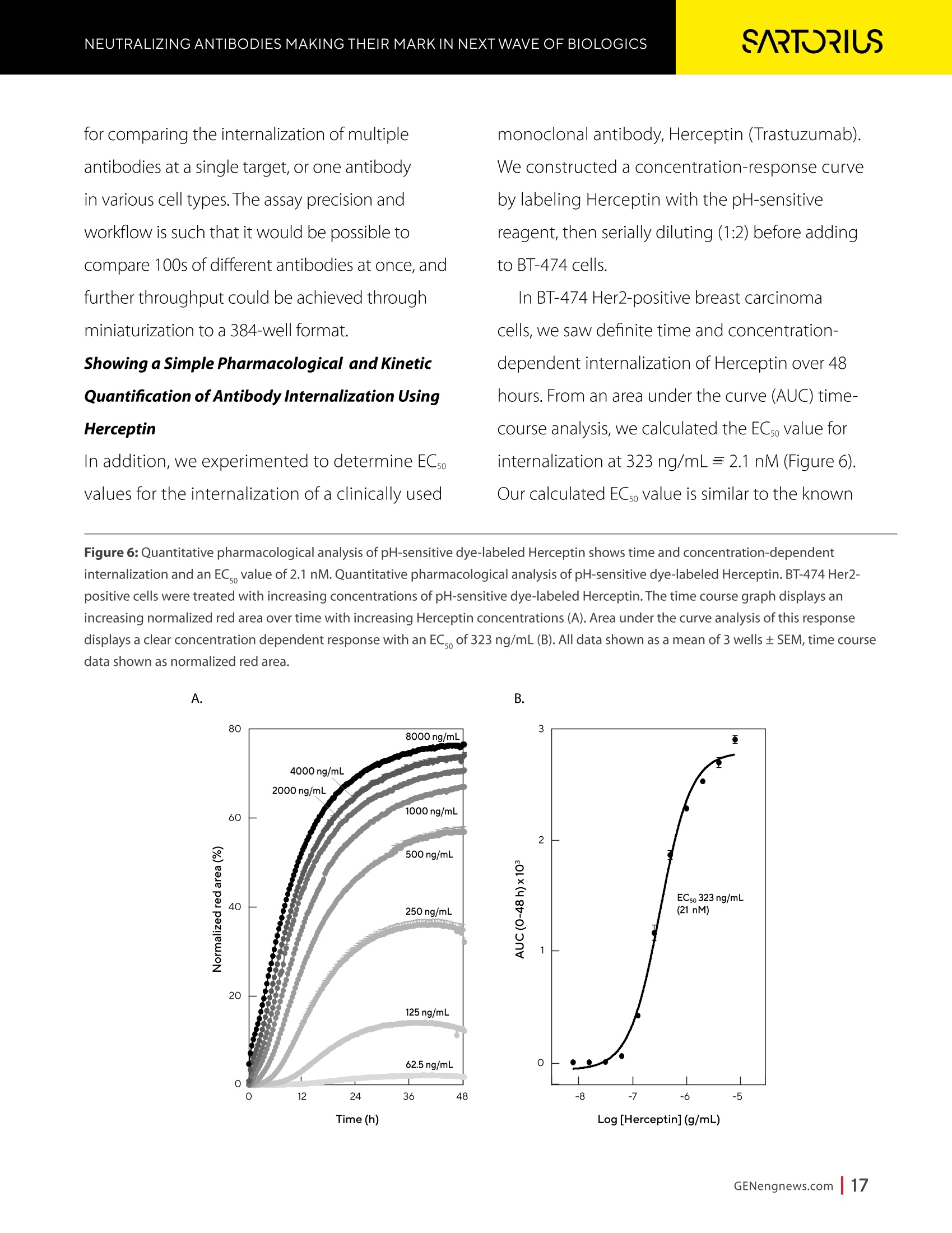
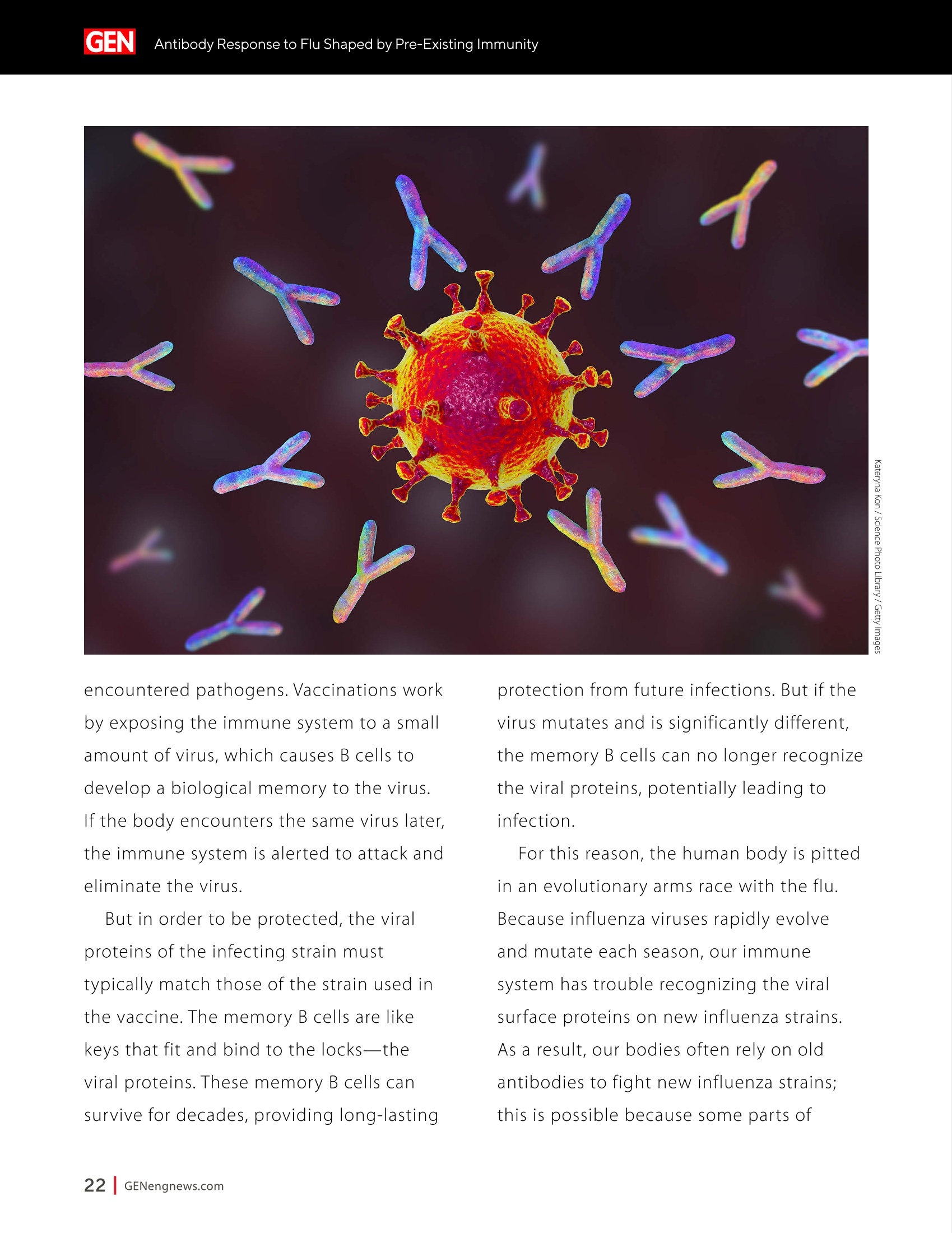
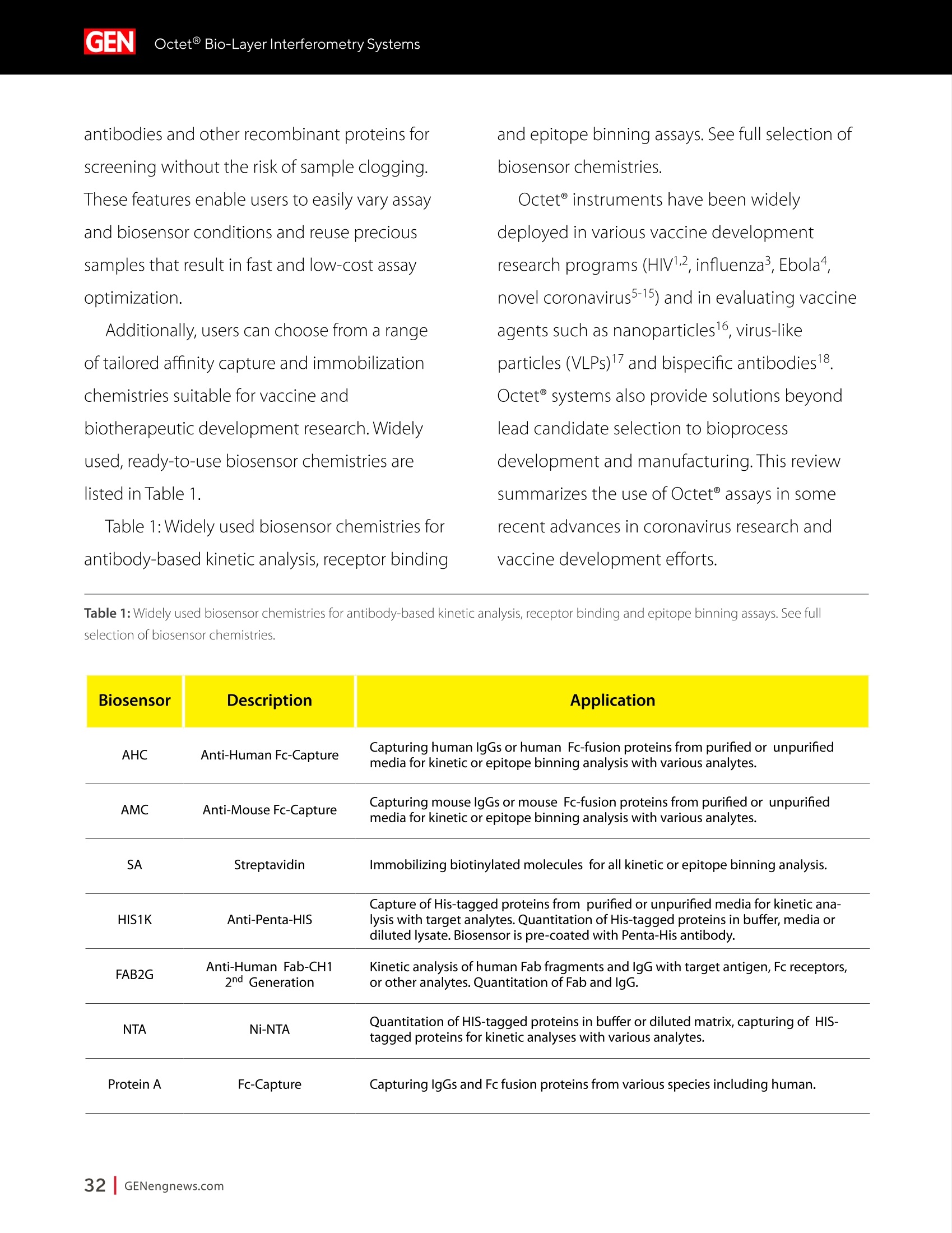
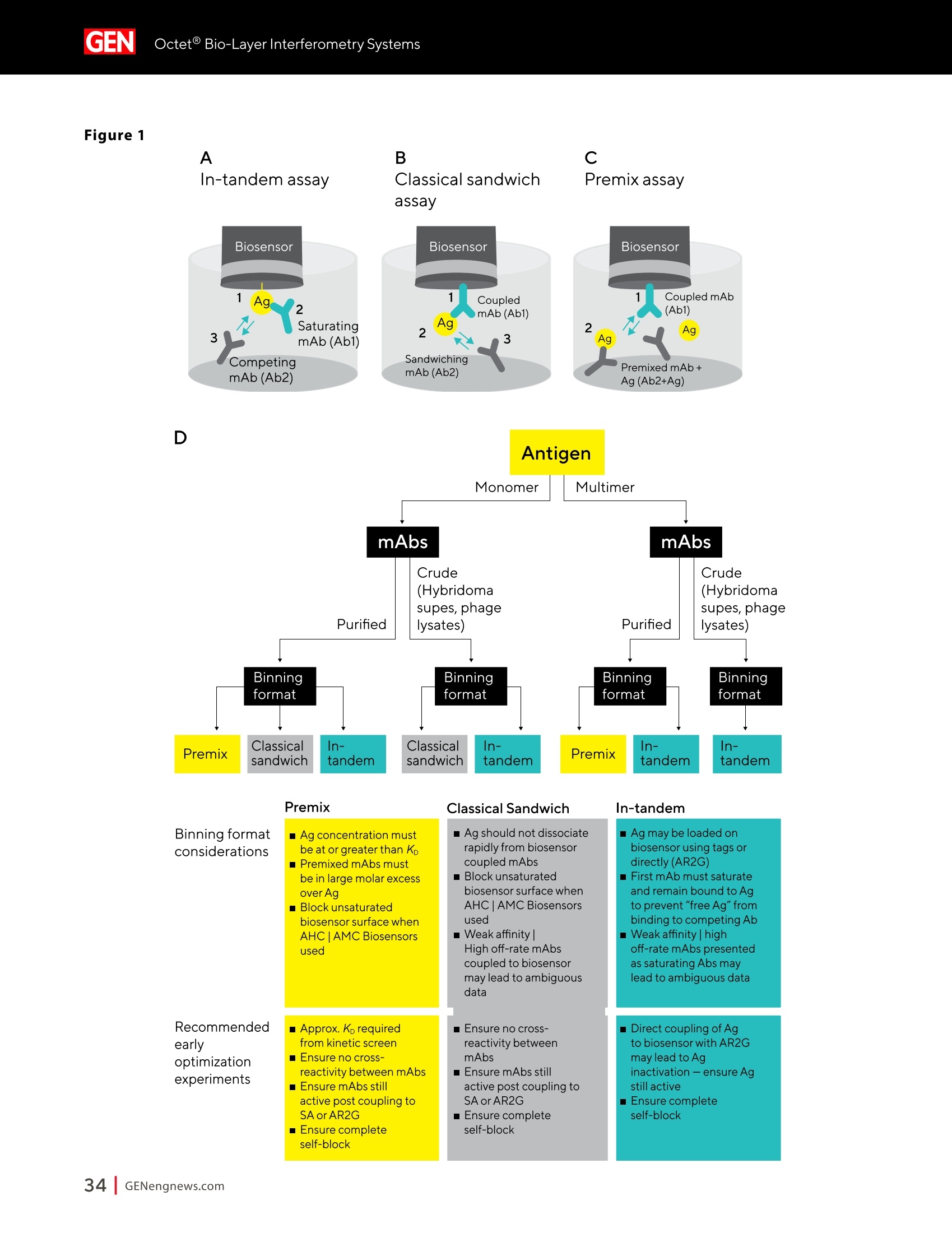
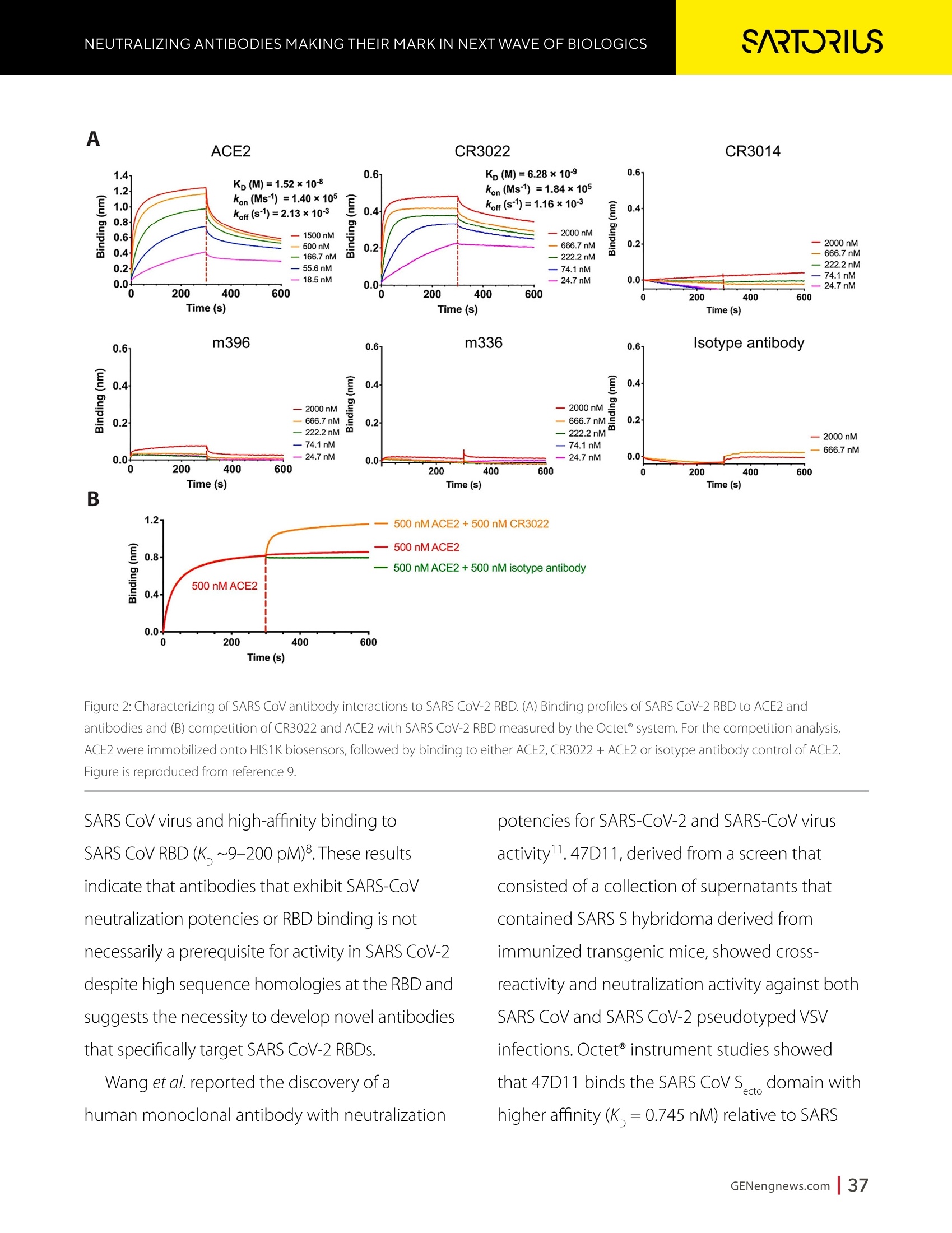
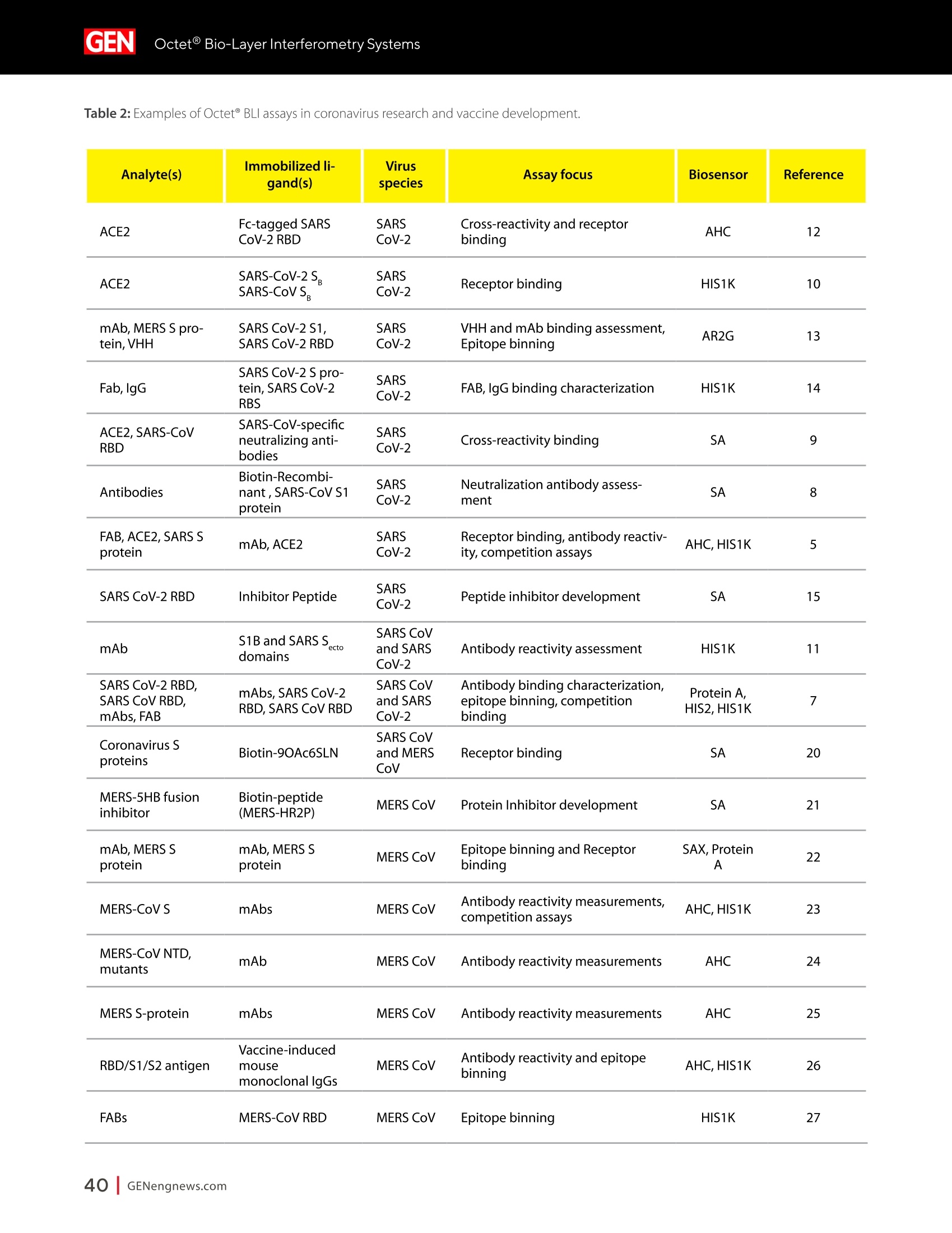
A SPONSORED PUBLICATION FROMSARTORIUSGenetic Engineering&Biotechnology News SARTORILSNEUTRALIZING ANTIBODIES MAKING THEIR MARK IN NEXTWAVE OF BIOLOGICS GEN Neutralizing AntibodiesMaking Their Markin Next Wave of Biologics SARTORILS Information-Rich Solutions toTransform Biologics Discovery Sartorius is transforming biologics discovery through innovation with ourgroundbreaking Octet,iQue° and Incucytecell & protein analysis platforms thatallow you to gain more insights and make better decisions. These unique, high capacity solutions provide information-rich data for tracking ofcomplex biological processes to generate deeper, more biologically relevantinsights, streamline workflows and accelerate biologics discovery anddevelopment. Simplifying Progress Learn more at: sartorius.com/biologics * 8 INTRODUCTION n the last couple of years, there has been heightened interestin the development of Monoclonal antibodies (mAbs) astherapeutics as they have shown great promise in the treatment ofserious or untreatable conditions such as cancer. This is partly due totheir highly specific targeting of antigens as well as their high efficacyand fewer side effects. Vaccines and effective antibody therapeuticswill play an integral role in keeping the on-going COVID-19 pandemicand emerging variants under control. Neutralizing Antibodies arean example of molecules that will play a critical role in vaccine andtherapeutics development as they can bind to a virus in a mannerthat blocks infection. Neutralizing Antibodies can be isolated fromplasma of convalescent patients and have become a major focus forresearchers around the globe in the race to a better understandingof the mechanisms of action (MOA) against the receptor-bindingdomains (RBD) of the SARS-CoV-2 Spike protein. In this collection we highlight the research efforts and relatedapplications utilized to study binding affinity, potency and targetspecificities of neutralizing antibodies against SARS-CoV-2 and theirpotential for future use against diseases caused by viruses from thesame coronavirus subgroup. The novel findings provide deeperinsights into the immune responses to infection, and help further theunderstanding of the interactions between the spike protein on theSARS-CoV-2 virus and its receptor on host cells. The interruption ofthis interaction has become the key target of COVID-19 therapeutics. Sartorius continues to support these early stage investigations byproviding innovative Incucyte@ live-cell analysis and iQue° advancedflow cytometry andOctet@label-free biomolecular interactionsanalysis platforms to provide new critical information, withconfidence, as quickly as possible. Learn more at— Monoclonal Antibody Discovery and DevelopmentSartorius Coronavirus-Neutralizing Human AntibodyDiscovered Antibody Internalization:Advanced FlowCytometry and Live-Cell Analysis Give RichInsights During Antibody Profiling Antibody Response to Flu Shaped byPre-Existing Immunity SARS-CoV-2Variants B.1.351 and B.1.1.7Show Resistance to Neutralizing Antibodies Synthetic Nanobodies,"Sybodies,"FoundThat Neutralize SARS-CoV-2 Octet@ Bio-Layer Interferometry Systems:Advancing Development of CoronavirusVaccine and Therapeutics Coronavirus-NeutralizingHuman Antibody Discovered esearchers at Utrecht University,ErasmusRMedical Center and Harbour BioMed(HBM) have identified a fully humanmonoclonal antibody that prevents SARS-CoV-2from infecting cultured cells. Discovery of the antibody, which also neutralizes the relatedSARS-CoVcoronavirus, represents an initial steptowards developing a fully human antibody totreat or prevent COVID-19, and also potentiallyfuture diseases caused by viruses from the samecoronavirus subgroup. "This discovery provides a strong foundationfor additional research to characterize thisantibody and begin development as a potentialCOVID-19 treatment"said Frank Grosveld, PhD, co-lead author on the study, AcademyProfessor of Cell Biology, Erasmus MedicalCenter, Rotterdam and Founding CSO at HarbourBioMed."The antibody used in this work is 'fullyhuman, allowing development to proceed morerapidly and reducing the potential for immune-related side effects." “Using this collection ofSARS-CoVantibodies,we identified an antibodythat also neutralizesinfection of SARS-CoV-2in cultured cells." -Frank Grosveld,PhD, Erasmus Medical Center, Rotterdam Grosveld and colleagues report on theantibody in Nature Communications, in a papertitled,"A human monoclonal antibody blockingSARS-CoV-2 infection." Both SARS-CoV-2 and the SARS-CoV virus thatemerged in 2002, belong to the Sarbecovirussubgenus of the Betacoronavirus family ofcoronaviruses.The two viruses crossed speciesbarriers from an animal reservoir, and can causelife-threatening respiratory illness in humans. ByMay 4th, 2020 there were more than 3.4 millionconfirmed cases of SARS-CoV-2 worldwide,and in excess of 230,000 deaths. The SARS-CoV strain caused ~8000 infections, with a lethality of10%. There are currently no approved targetedtherapeutics are available for COVID-19, thedisease caused by SARS-CoV-2. Monoclonal antibodies targeting "vulnerablesites"on viral surface proteins are increasinglyrecognized as a promising class of drugsagainst infectious diseases, and have showntherapeutic efficacy for a number of viruses,the authors wrote. Coronavirus-neutralizingantibodies primarily target the trimeric spike (S)glycoproteins on the coronavirus surface thatmediate entry into host cells. The S protein hastwo functional subunits. The S1 subunit,whichis composed of four core domains, S1 throughto S1, mediates attachment to the host cell. TheS2 domain mediates fusion of the viral and cellmembranes. The spike proteins of SARS-CoV-2 andSARS-CoV share 77.5% identical amino acidsequence, and are structurally very similar, theinvestigators continued.They commonly bind thehuman angiotensin converting enzyme 2 (ACE2)protein as the host receptor."Potent neutralizingantibodies often target the receptor interactionsite in S1, disabling receptor interactions"theauthors continued. For their reported antibody discovery effort,Grosveld and colleagues built on work that thegroups had carried out on antibodies targetingSARS-CoV, explained co-lead author Berend-Jan Bosch, Associate Professor, research leader atUtrecht University."Using this collection ofSARS-CoV antibodies, we identified an antibodythat also neutralizes infection of SARS-CoV-2 incultured cells.The human antibody identified,47D11, was generated using Harbour BioMed'sH2L2 transgenic mouse technology. 47D11 was shown to bind to cells expressingthe full-length spike proteins of both SARS-CoVand SARS-CoV-2, and potently inhibited viralinfection of cultured cells. Tests showed thatthe antibody targeted the S1, receptor-bindingdomain (RBD) of the spike proteins of bothviruses. The fact that the antibody is cross-reactiveindicates that it likely targets the conservedcore structure of the S1, RBD, the investigatorssuggested."Such a neutralizing antibody haspotential to alter the course of infection in theinfected host, support virus clearance or protectan uninfected individual that is exposed to thevirus,Bosch stated. Interestingly, the team's results suggested that47D11 neutralizes SARS-CoV and SARS-CoV-2through "a yet unknown mechanism"that isdifferent from receptor-binding interference."Alternative mechanisms of coronavirus neutralization by RBD-targeting antibodieshave been reported including spike inactivationthrough antibody-induced destabilization of itsprefusion structure, which may also apply for47D11, the team noted. "In conclusion, this is the first report of a(human) monoclonal antibody that neutralizesSARS-CoV-2"they concluded."This antibodywill be useful for development of antigendetection tests and serological assays targetingSARS-CoV-2... this antibody-either alone orin combination—offers the potential to preventand/or treat COVID-19, and possibly also otherfuture emerging diseases in humans caused byviruses from the Sarbecovirus subgenus" "This is groundbreaking research,"saidJingsong Wang,PhD, founder, Chairman& Chief Executive Officer of HBM."Muchmore work is needed to assess whether thisantibody can protect or reduce the severityof disease in humans. We expect to advancedevelopment of the antibody with partners.We believe our technology can contributeto addressing this most urgent public healthneed and we are pursuing several otherresearch avenues. AntibodyInternalization: Advanced FlowCytometry and Live-Cell Analysis Give RichInsights During Antibody Profiling CLARE SZYBUT1*CAROLINE WELDON2*NICOLABEVAN1*LORI KING 1. Essen BioScience, Ltd., Part of the Sartorius Group, Units 2 & 3 The Quadrant, Newark Close, Royston Hertfordshire SG8 5HL, UK 2. 2. Essen BioScience, Inc., Part of the Sartorius Group, 5700 Pasadena Ave. NE, Albuquerque,NM 87113 USA Introduction he natural characteristics of antibodies, such as high binding affinity, specificityto a wide variety of targets, andgood stability, make them ideal therapeuticcandidates for many diseases.Monoclonalantibodies (mAbs), in particular, deliver promisingtherapeutic results in several different diseaseareas, such as autoimmunity, oncology, andchronic inflammation. Researchers'abilities toimprove the breadth of antibodies have beenaided by innovative technologies for antibodydiscovery, for instance, through humanization ofmouse antibodies and phage display. However,advanced antibody design techniques createthe need for new screening methods so thatlead candidates can be quickly and effectivelyidentified as early in the development process aspossible. Part of an effective screening strategy is toidentify the desired therapeutic goal for your candidates. For instance, binding and rapidinternalization are desirable properties forantibody-drug conjugates (ADCs), as cells mustbe selectively targeted and killed via deliveryof a cytotoxic payload. In contrast, if the goal isto induceantibody-dependent cell-mediatedcytotoxicity (ADCC), the antibody must remainbound to the cell surface, rather than beinginternalized, in order to activate an immuneresponse. When designing ADCs, one key attribute topredicting efficacy is the antibody internalization(ABl) rate and the associated kinetics. Theseinternalization kinetics are influenced by factorssuch as the epitope on the target antigen, affinityof the ADC-antigen interaction, and intracellulartrafficking. Evaluating for these factors is criticalthroughout the antibody screening pathway(Figure 1); however, here, we concentrate onevaluating antibodies'ABI during functionalprofiling via rate comparisons during the Figure 1: Inclusion of ABl in the screening pathway for antibodies. Selecting for ABl throughout the antibody screening pathway, whether during functional profiling and rate comparisons or mechanistic studies. development process, as well as multiplexedmechanistic studies later in the screening process. Traditionally, antibody screeningworkflows have required labor intensive, timeconsuming, end-point-onlymethods, suchas FACS, ELISA, or microscopy. One solutionto address these screening challenges is touse advanced screening methods that offermaximum insight through high-content data.Herein, we demonstrate an advanced antibodyscreening method that focuses on speed and insight by using a novel, pH-sensitive reagentforcharacterizing ABl via both advanced flowcytometry and live-cell imaging. Challenges with TraditionalAntibody Screening Workflows Monoclonal antibodies (mAbs) are large(about 150 kDa), complex biologic moleculesthat require post-translational modificationsfor their activity (Chames et al., 2009). Therefore, researchers face several challenges when engineering and producing mAbsas therapeutics. Engineering antibodies tooptimize their biological potencies duringthe discovery phase can address many ofthese challenges. However, these attempts tooptimize one attribute can have profound andunintended consequences on other antibodyattributes. For instance, optimizing an antibody'sspecificity may negatively affect activity(Tiller and Tessier, 2015). One way of simultaneously optimizingmultiple antibody properties is by usingmutagenesis to produce large screeninglibraries. However, large screening librariesnecessitate a thorough in vitro,high-throughput screening method to quicklyidentify the most suitable drug candidatesfor further development early in theirdiscovery process. Workflows for antibody screening commonlyinclude fluorescence-activated cell sorting(FACS),enzyme-linked immunosorbent assays(ELISA), and microscopy (such as confocal)techniques. Yet, these in vitro antibody screeningmethods have several drawbacks: (1) they canbe labor intensive with limited throughput,(2) they do not allow direct, head-to-headcomparisons of antibodies, and (3) they needlarge amounts of reagents. For instance, even though FACS can be usedto sort hundreds of thousands of cells based on their fluorescence properties, it cannot be usedto perform high-throughput,time-dependentstudies on individual cells because the cellsmust be sorted into single colonies before anyfurther analysis can be performed (Doerneret al.,2014). ELISA, on the other hand, is adaptable tohigh-throughput screening (Saeed et al., 2017),and thus has historically been used to screenhybridomas and other libraries for antibodybinding to each of its targets. ELISAs areperformed by coating a single target antigenonto the wells of assay plates, followed by theaddition of individual samples from an antibodylibrary (for instance, from hybridoma or phagedisplay).Antibodies that bind to the immobilizedantigen are detected by a color change due to anindirect enzyme/substrate reaction. In addition to its adaptability to high-throughput screening, ELISA is rapid, consistent,and relatively easy to analyze (Saeed et al.,2017).However, ELISA has several disadvantagesthat can limit its successful use in a modernantibody screening lab. First, primary screensthat test binding to a single antigen often requiresubsequent secondary and sometimes eventertiary screens with control antigens to confirmtheir specificity and cross-reactivity. Second, ELISAis not the best method for screening antibodiesthat bind to cell surface antigens because theseantigens are extracted from the cell membrane and purified before adsorbing to the plastic ELISAplate. Extracting antigens from the membraneoften leads to disruption of conformationalepitopes that can be important targets fortherapeutic antibodies. Finally, to minimizebackground signal, ELISA requires multiplewash steps to remove unbound antibodies anddetection reagents, resulting in long, labor-intensive screening workflows. Also,although ELISA can provide data on theimmunoglobulin G (lgG)titer, it offers no reflec-tion on how the therapeutic antibody candi-dates affect the health of the test cells. i.e.howquickly or efficiently they induce death.Thus,candidates that appear to be productive basedon ELISA screening may be carried forward intothe next step of the production process eventhough they are unideal candidates in terms oftunction or vigour. Microscopy techniques, in contrast to FACS,are useful for single-cell analysis, as well asfor such localization and temporal studies asantibody internalization and live-cell imagingfor monitoring individual cell behavior.Microscopy techniques, however, have limitedthroughput due to data acquisition time, andthey have limited multiplexing capabilities.Thus, for antibody discovery, some othermethod, preferably high-throughput, isgenerally used for the initial selection andscreening of large libraries, followed by a more thorough functional analysis of a smaller setof antibody candidates using microscopy(Doerner et al., 2014). Like FACS, microscopy techniques also requirelabeling each antibody with a fluorescent tag,which must be separated from the free label viaa column or wash step because analysis requiresrobust isolation of internalized antibodiesfrom those outside the cells. To aid isolationof the positive signal, researchers often resortto perturbing techniques, such as washingcells, using blocking dyes, and reducing thetemperature to slow cellular activity. However,cells can be lost during washing steps, and theassociated reductions in temperature perturb thecellular environment. A further drawback to almost all thetechniques used for antibody screening isthat they only enable end-point analysis,which means that multiple experiments arerequired to follow an antibody attribute, such asinternalization, over time. The best approach to address all thelimitations of traditional antibody screeningis to combine data from advanced screeningmethods; using advanced methods,researchers can choose their candidatesbased on a thorough evaluation of all relevantcharacteristics, such as lgG titer, cell health,internalization,and their associated kineticsearly in their discovery process. AntibodyInternalization Reagent Principles Figure 2: The pH-sensitive fluorescent probe principle. A novel pH-sensitive fluorescent probe enables one-step,no-wash labelingof isotype-matched antibodies. A fluorescent signal is generated as internalized antibody is processed into the acidic endosome andlysosomepathway. A Simple Way of Labeling Antibodiesfor Addressing Challenges withFACS, ELISA, and Microscopy One of the challenges presented above withFACS,ELISA, and microscopy techniques isthe requirement for wash steps to minimizebackground signal. Among other undesirableeffects, such as increased time to results, thisneed for wash steps also increases reagentrequirements (and cost) and makes cell lossinevitable. However, an advanced method forlabeling cells could reduce reagent requirements,as well as simplify protocols when analyzingantibody internalization. ABI Assays Made Simple, With No-Wash Labelingand Low Reagent Requirements via a Novel, Ph-Sensitive Reagent The key to this simple, no-wash protocol is anovel reagent composed of Fc-region targeting Fab fragments conjugated to a pH-sensitivefluorescent probe (Nath et al., 2016). This typeof novel reagent enables a generic, one-step,no-wash labeling protocol for all isotype-matched, Fc-containing test antibodies whenoptimized for use on specific instruments.Figure 2 shows how this reagent works: labeledantibodies are added to cells, and a fluorogenicsignal is produced as the Fab-Ab complex isinternalized and processed via acidic (pH 4.5-5.5)lysosomes and endosomes. Antibodies of interest are quickly andeffectively labeled, with low reagentrequirements, by incubating in growth mediawith this novel, pH-sensitive dye (Figure 3).Cells are then added to 384-well plates, alongwith the dye-conjugated antibodies, andincubated again. This reagent, when used withan advanced flow cytometry platform, provides Multiplexed Viability/Encoding Antibody Internalization Reagent Workflow a comprehensive, integrated solution for rapidprofiling of antibody internalization andother critical antibody attributes using smallsample volumes in 96- or even 384-well plateformats (Riedl et al.,2016). Much of the dataacquisition and analysis, including generationof serial dilution curves and ECso calculations, isautomated with the help of advanced softwarepackages, for example, as included in the iQueadvanced flow cytometry platform. Functional Profiling forComprehensive Cell and AntibodyCharacterization Early in theProcess with Multiplex Analysis Combining the above-described pH-sensitive dyewith other reagents in one assay on an advancedflow cytometry platform enables simultaneousanalysis of a variety of cell and antibody characteristics within the same workflow. Forinstance, you can measure cell viability using amembrane integrity dye to assess general cellhealth, as well as cell death due to cargo delivery,when optimizing ADCs. Or you can characterizecell specificity using encoding dyes (for cell lines)or directly conjugated fluorescent antibodies(for complex cell models with a variety of celltypes).Other reagents, such as those for assessingcytokine release, are also available for moredetailed antibody assessments on the sample.Therefore, analysis with an advanced flowcytometry platform can be optimized to deliverrich content very quickly while only using a lowsample volume. Sartorius produces such a pH-sensitivereagent for use on ouriQue" advancedflow cytometry platform. The combinationof non-perturbing and validated reagents A. -e-CD19 -CD20 CD79b CD71 +- CD19 CD20-CD79b CD71 -CD19 -0CD20-CD79b CD71 →mlgG1 -CD22 CD3 + mlgG1 CD22 CD3 +mlgG1 CD22 CD3 100000.0 60000.0oc oc cc 100000.0 o 80000.0 一工50000.0co一c 80000.0 60000.0 40000.0 60000.0 30000.0 40000.0 40000.0 20000.0 20000.0 20000.0 -零 10000.0 0 2 3 2 3 2 3 log(Concentration)pg/mL log(Concentration)pg/mL log(Concentration)pg/mL B. Jurkat Ramos Figure 4: Serial dilution curves for internalization-labeled antibodies with a top concentration of 1 mg/mL in different cell types aftera three-hour incubation.Multiplexed positive and negative cell lines may be used in an ABl assay to generate high-content data inone assay. Median fluorescent intensity (MFI) for Internalization Reagent-labeled antibodies after three hours. A serial dilution ofeach antibody with a top concentration of 1 mg/mL was prepared and incubated with encoded Jurkat, Raji, and Ramos cells in thesame well. Jurkat cells (a T lymphocyte cell line) show a concentration-dependent increase in internalization of anti-CD3, but not thetwo B cell markers. Conversely, Raji cells show a concentration-dependent increase in internalization of anti-CD19 and anti-CD22,but not anti-CD3. Ramos cells show a concentration-dependent increase in internalization of anti-CD79b, an ADC drug target fornon-Hodgkin's lymphoma. for multiplexing, no-wash protocols,high-throughput capabilities, flexibility for arobotic interface, and integrated Forecytsoftware for multiparametric data analysis and visualization expedite the process of screeningdrug candidates for potential efficacy andtoxicity to accelerate antibody discovery anddevelopment. To demonstrate the multiplexingcapabilities of this novel pH-sensitive dye onthe iQue"platform, we used Ramos and Rajicells stained with two intensities of violetencoding dye, combined with unstainedJurkat cells. We incubated this forthree hourswith a serial dilution of dye-conjugatedspecificity antibodies (isotype-matchedanti-CD3 as a T cell marker,anti-CD19,anti-CD20,anti-CD22, or anti-CD79b as B cellmarkers,anti-CD71 as a positive control,andIgG as a negative control), then added a cellmembrane integrity dye before acquiring datafrom the plate. Using this strategy, we wereable to identify viable cells, then spectrallyseparate Ramos, Raji, and Jurkat cells. We then assessed antibody internalizationfor each cell line. We generated series dilutioncurves for the specificity markers of each celltype (Figure4A). As expected, Jurkat cells showedinternalization of anti-CD3, but not anti-CD19or anti-CD22, whereas the Raji cells internalizedanti-CD19 and anti-CD22, but not anti-CD3. OnlyRamos cells showed a concentration-dependentincrease in internalization of anti-CD79b, an ADCdrug target for non-Hodgkin's lymphoma. In thethree-hour assay time frame, we did not observeanti-CD20 internalization,but we did see anincrease in the two B cell lines by 24 hours (datanot shown).Importantly, we saw little differencewhen we assessed the cells for internalization alone or when we mixed them. This resultshows that multiplexing positive and negativecell lines does not interfere with the antibodyinternalization assay. Compared to performing a series of singleplexassays, a multiplexed assay approach enablesyou to analyze multiple readouts (internalization,viability, cell type) from a single well, decreasingthe number of tests needed to perform acomprehensive functional characterization of theAb candidate. Full Concentration Profiling withLive-Cell Imaging and Analysis A pH-sensitive dye-coupled antibody fragmentdesigned according to the same principle asthat used above (Figure 2) can also be optimizedand used in other instruments. For instance.using this pH-sensitive reagent in a real-time,live-cell imaging system such as the IncucyteLive-Cell Analysis System, allows visualization andautomatic quantification of the full time-course ofABI. This combination of reagent and platform thusprovides a simple method for directly profiling andcomparing ABl for a large number of antibodies(10-100s at a time in a miniaturized format). To demonstrate the power of the pH-sensitivedye approach for high-throughput antibodyinternalization assays in a real-time, live-cellanalysis system, we performed a head-to-headcomparison of the internalization properties of Figure 5: Screening test of Abs for internalization. The pH-sensitive reagent is suitable for high-throughput antibody internalizationassays in a real-time, live-cell analysis system. six different commercially available anti-CD71antibodies into HT1080 fibrosarcoma cells. Welabeled the anti-CD71 with our pH-sensitivereagent before adding to cells in 96-well plates.We then captured the internalization signal in alive-cell analysis system every 30 minutes over12 hours using a 10X magnification. The plate view in Figure 5A shows clearpositive and negative control responsesin column 11 and 12, with concentration-dependent responses for each antibody acrossthe two plates (antibodies 1a and 1b are the same clone from two different sources). We found that three antibodies (Ab1a,Ab2,and Ab1b) gave internalization signals that wedetected at low concentrations (< 0.05 ug/ml)(Figure 5).Reassuringly, Ab1a and Ab1b gavesimilar internalization responses.Antibodies3, 4, and 5 were internalized more weakly andonly at higher concentrations. From the controlresponses, we calculated a mean Z'value of0.82 (two plates: 0.75, 0.87), indicating highrobustness for this microplate assay. These data show our method is suitable for comparing the internalization of multipleantibodies at a single target, or one antibodyin various cell types. The assay precision andworkflow is such that it would be possible tocompare 100s of different antibodies at once,andfurther throughput could be achieved throughminiaturization to a 384-well format. Showing a Simple Pharmacological and KineticQuantification of Antibody Internalization UsingHerceptin In addition, we experimented to determine ECsovalues for the internalization of a clinically used monoclonal antibody, Herceptin (Trastuzumab).We constructed a concentration-response curveby labeling Herceptin with the pH-sensitivereagent, then serially diluting (1:2) before addingto BT-474 cells. In BT-474 Her2-positive breast carcinomacells, we saw definite time and concentration-dependent internalization of Herceptin over 48hours. From an area under the curve (AUC) time-course analysis, we calculated the ECso value forinternalization at 323 ng/mL=2.1 nM (Figure 6).Our calculated ECso value is similar to the known Figure 6:Quantitative pharmacological analysis of pH-sensitive dye-labeled Herceptin shows time and concentration-dependentinternalization and an EC_value of 2.1 nM. Quantitative pharmacological analysis of pH-sensitive dye-labeled Herceptin. BT-474 Her2-positive cells were treated with increasing concentrations of pH-sensitive dye-labeled Herceptin. The time course graph displays anincreasing normalized red area over time with increasing Herceptin concentrations (A). Area under the curve analysis of this responsedisplays a clear concentration dependent response with an EC_of 323 ng/mL (B). All data shown as a mean of 3 wells ± SEM, time coursedata shown as normalized red area. A. B. Time(h) Log [Herceptin] (g/mL) KD value for Herceptin for its target receptor(approximately 5nM). Speed and Insight throughAdvanced Antibody Screening Quick and accurate identification of suitabledrug candidates is key to the development oftherapeutic antibodies. ABl is an essential partof the selection criteria for ADC candidates. Itcan be used for functional profiling and ratecomparisons, as well as mechanistic studieswhen coupled with additional multiplexedreadouts, thus reducing the time required forlead generation. To illustrate the capabilities ofadvanced antibody screening, we have describedour solution for efficiently interrogating libraries ofcandidates early in the screening process. For comprehensive cell and antibodycharacterization, we used our novel, pH-sensitivereagent and the iQue advanced flow cytometryplatform.This combination is best for screeninaand early full concentration profiling because itenables simultaneous analysis of a variety of celland antibody characteristics within the sameworkflow. For quantitative, pharmacologicalanalysis and direct,head-to-head comparisons ofABl, we used a version of our novel, pH-sensitivereagent and the Incucyte live-cell analysis system.This combination is useful for further functionalprofiling that requires spatial and temporalresolution. This advanced antibody screening process is a simple, fast, and insightful way toidentify candidates that meet your therapeuticgoals early in the drug discovery process. Combining Advanced Flow Cytometryand Live-CellImaging and Analysisfor Complete Antibody Profiling:A Real-World Screening StrategyEmploying Our pH-Sensitive Dyeand Instrument Platforms Researchers at LifeArc used the iQue platformand Incucyte Live-Cell Analysis System as partof their strategy to develop a new ADC targetingneuroblastoma.Neuroblastoma is a rare cancerthat nevertheless is the most common extra-cranial solid tumor in children, with a 5-yearsurvival rate of 50% for patients with high-riskdisease. The researchers found they were ableto use these systems in combination, not onlyfor screening, but also for assay development,lead candidate profiling, and characterization.They found that the iQue platform offered fast,high-content analysis, while the Incucyte live-cell analysis system offered kinetic,image-basedanalysis. Combining data from the two systemsgave them a complete antibody profile. In brief, the researchers identified anaplasticlymphoma kinase (ALK) as a target for theirtherapeutic approach because level of ALKexpression correlates with disease stage andALK antibodies show surface expression in patient samples. In collaboration withcolleagues at Mt. Sinai, researchers at LifeArcproduced 1,152 candidate hybridoma clonesthat they were able to narrow to 53 candidatesthat ELISA and flow cytometry showed boundto ALK. Using the advanced flow cytometrycapabilities of the iQueplatform, theresearchers were able to further narrow this to20 candidates that bound ALK at the surfaceof cells, since this is a critical attribute of themechanism of action of ADCs. Ofparticular interest, these researchers usedinternalization assays on both the iQue andIncucyte cell analysis platforms to further narrowtheir lead candidates to two. Critically, theygained kinetic imaging data and high-contentanalysis using multiplexed cell viability assaysearly in their ADC discovery process. Using advanced screening methods that incorporate anovel, innovative reagent for characterizing ABIvia both advanced flow cytometry and live-cellimaging, they gained maximum insight throughhigh-content data and were able to make criticaldecisions early in their process, thus saving time,material, and money. References ( T iller KE and Tessier PM. Advances in antibody design. Annu Rev Biomed Eng, Feb 5; 17: 19 1 -216(2015) PMI D : 262 7 4600 ) ( Doerner A, et al. Therapeutic antibody engineering by high efficiency cell screening. FEBS Lett, Jan 21;588(2);278-87 (20 1 4) PMID: 24291259 ) ( Nath N, et al. Homogeneous plate based antibody internalization assay using pH sensor fluorescent dye.JImmun Meth, Feb 3; 431:11-21 (2016) PMID: 26851520 ) ( Reidl T, et al.High-Throughput Screening for Internalizing Antibodiesby Homogeneous Fluorescence Imaging of a pH-Activated Probe.J Biomol Screen, Jan; 21(1):12 - 23(2016) PMID: 26518032 ) Chames P,et al. Therapeutic antibodies: successes, limitations andhopes for the future. BrJPharmaco, May; 157(2): 220-33 (2009) PMID:19459844 Saeed A, et al. Antibody Engineering for Pursuing a Healthy Future.Front Microbiol, Mar; 8(495) (2017) PMID: 28400756 Antibody Response to FluShaped by Pre-Existing Immunity eceiving the seasonal flu vaccine each year,Rin addition to seasonal infections, exposespeople to a lifetime of building up immuneresponses to influenza antigens. Yet, it remainsunclear whether infection and vaccinationinduce distinct influenza-specific immunologicalmemory. A team led by researchers at theUniversity of Chicago compared antibodiesproduced by individuals after influenza infection or vaccination.The authors found that aperson's antibody response to influenza virusesis dramatically shaped by their pre-existingimmunity, and that the quality of this responsediffers in individuals who are vaccinated ornaturally infected. Their results highlight theimportance of receiving the annual flu vaccine toinduce the most protective immune response. The study is published in Science Translational Medicine, in an article titled,"Preexisting immunity shapes distinctantibody landscapes after influenza virusinfection and vaccination in humans. The researchers compared antibodiesproduced by individuals after influenzainfection or vaccination. They found thatinfection-induced antibodies reacted tonon-neutralizing epitopes of influenza virus,whereas vaccination-induced antibodiesreacted to neutralizing epitopes. The team found that most of the initialantibodies stimulated after both influenzainfections and influenza vaccinationscame from old B cells, indicating theimmune system's memory plays a majorrole in how the body responds early onto a viral infection. These antibodiesdisplayed higher reactivity toward strainsof influenza that circulated during anindividual's childhood compared to morerecent strains. "Most interestingly, we found that peoplewho were actively sick with influenza hadold antibodies that predominantly targetedparts of the virus that don't change—but those antibodies specifically targetednon-neutralizing sites" said Haley Dugan,co-first author of the study and a PhDcandidate in immunology.“When we testedthese same antibodies in mice, they weren'table to protect them from being infectedwith influenza." In contrast, the researchers found that "This study provides a majorframework forunderstandinghow pre-existing immunityshapes protective antibodyresponses to influenza inhumans, -Patrick Wilson, PhD,University of Chicago influenza vaccinations boost antibodiesthat tended to target conserved yetneutralizing regions of the virus, whichsuggests vaccinations can draw uponpre-existing immunity to prompt moreprotective responses. Vaccinated individualsalso generated many antibodies thattargeted new and mutated regions on thevirus, suggesting these vaccine-inducedantibodies are more adaptable. Immune system memory ensures arapid and specific response to previously encountered pathogens. Vaccinations workby exposing the immune system to a smallamount of virus, which causes B cells todevelop a biological memory to the virus.If the body encounters the same virus later,the immune system is alerted to attack andeliminate the virus. But in order to be protected, the viralproteins of the infecting strain musttypically match those of the strain used inthe vaccine. The memory B cells are likekeys that fit and bind to the locks-theviral proteins. These memory B cells cansurvive for decades, providing long-lasting protection from future infections. But if thevirus mutates and is significantly different,the memory B cells can no longer recognizethe viral proteins, potentially leading toinfection. For this reason, the human body is pittedin an evolutionary arms race with the flu.Because influenza viruses rapidly evolveand mutate each season, our immunesystem has trouble recognizing the viralsurface proteins on new influenza strains.As a result, our bodies often rely on oldantibodies to fight new influenza strains;this is possible because some parts of the influenza virus that are critical toits structure or function do not change,remaining familiar to our immune system. Researchers now understand thatspecific structural and functional partsof the influenza virus that do not changeare better for antibodies to target thanothers.Antibodies that bind to one ofthese neutralizing sites are able to preventinfection, while antibodies that targetnon-neutralizing sites often cannot. Scientistsbelieve a person's age, history of exposure tothe influenza virus, and type of exposure-either through infection or vaccination-all shape whether their immune systemantibodies target neutralizing ornon-neutralizing sites on a virus. In the study, scientists sought toaddress a major knowledge gap: Whichconserved viral sites are preferentiallytargeted following natural infectionversus vaccination in people, and howdoes pre-existing immunity play a role inshaping the landscape of neutralizing andnon-neutralizing antibodies? "For people who have caught the flu, their pre-existing immunity may makethem susceptible to infection or increasethe severity of their influenza symptomsif their antibodies are targeting 'bad' ornon-neutralizing viral sites," said co-firstauthor and immunology postdoctoral fellowJenna Guthmiller, PhD. By contrast, vaccination largely inducesneutralizing and protective antibodies, oldand new, highlighting the importance ofreceiving the seasonal influenza vaccine. "This study provides a major frameworkfor understanding how pre-existingimmunity shapes protective antibodyresponses to influenza in humans,said Patrick Wilson, PhD, a professor ofimmunology and lead author of the study."We need more studies to determinewhether the targeting of specificneutralizing and non-neutralizing viral sitesdirectly impacts a person's likelihood ofbecoming ill." The researchers are now examining howearly exposure to the influenza virus inchildren shapes their immune response laterin life as a follow-up to this work. SARS-CoV-2Variants B.1.351and B.1.1.7Show Resistance toNeutralizing Antibodies ach passing week bringsF:more questions about theSARS-CoV-2 variants. One of the biggest questions has been whetherthe emerging variants will responddifferently to neutralizing antibodiesmade by people who have been infected.This point is important to understand,as it may alter the ability of our currentarsenal oftherapeutics and vaccines tocombat the infection. Now, new research from the labs at theAaron Diamond AIDS Research Center at theColumbia University College of Physiciansand Surgeons, suggests that two of thevariants,the B.1.351 and B.1.1.7 SARS-CoV-2variants (first detected in South Africa andthe U.K., respectively),show increasedresistance to antibody neutralization inlaboratory experiments. The findingssuggest that current antibody therapies andvaccines may be less effective against somevariants of the virus. This work is published in Nature, in the paper,"Antibody Resistance of SARS-CoV-2Variants B.1.351 and B.1.1.7" A preprint ofthe study was first posted to BioRxiv onJanuary 26,2021. Monoclonal antibodies, which targetspecific sites on the SARS-CoV-2 virus,arebeing used in hospitals to treat COVID-19.However, these therapeutics were designedto work against the initial variant ofSARS-CoV-2, which emerged in 2019. David Ho, MD, professor of microbiology& immunology and director, Aaron DiamondAIDS Research Center, and colleagues,assessed the ability of 30 monoclonalantibodies, along with plasma from 20patients who recovered from COVID-19and sera from 22 people who have beenvaccinated,to neutralize the B.1.351 andB.1.1.7 variants of SARS-CoV-2. The authors found that the B.1.1.7variant was resistant to neutralization bymonoclonal antibodies that target theN-terminal domain of the spike protein andwas relatively resistant to some antibodiesthat target the receptor-binding domain.However, it was not resistant to plasma frompatients who had recovered from COVID-19and sera from individuals who werevaccinated, and the authors suggest thatthis variant will not have a marked impacton current therapies or vaccines. However, in addition to resistance to neutralization by antibodies to theN-terminal domain, the B.1.351 variantwas found to be resistant to a group ofmonoclonal antibodies that is currently usedin therapies that target the receptor-bindingmotif of the spike protein, which wasprimarily attributed to the E484K mutation.The neutralizing activity of plasma frompatients who had recovered from COVID-19and sera from people who had beenvaccinated was reduced by approximately9- and 10-12-fold, respectively, against thisvariant. The study's predictions are beingborne out with the first reported resultsof the Novavax vaccine, said Ho. "Theapproximately 2-fold loss of neutralizingactivity against the U.K. variant is unlikelyto have an adverse impact due to the largecushion' of residual neutralizing antibodyactivity," Ho said, "and we see that reflected Novaris reported that the vaccine was nearly90% effective in the company's U.K. trial, butonly 49.4% effective in its South Africa trial,where most cases of COVID-19 are caused bythe B.1.351 variant. in the Novavax results where the vaccinewas 85.6% effective against the U.K. variant." Data from Ho's study about the loss inneutralizing activity against the South Africavariant are more worrisome."The drop inneutralizing activity against the South Africavariant is appreciable, and we're now seeing,based on the Novavax results, that this iscausing a reduction in protective efficacy,"Ho said. The company reported on January28 that the vaccine was nearly 90% effectivein the company's U.K. trial, but only 49.4% effective in its South Africa trial, where mostcases of COVID-19 are caused by theB.1.351 variant. The new study did not examine the morerecent variant found in Brazil (B.1.1.28) butgiven the similar spike mutations betweenthe Brazil and South Africa variants, Ho saidthe Brazil variant should behave similarly tothe South Africa variant. They argue that SARS-CoV-2 is mutatingin a direction that may cause it to evadecurrent interventions that are directedagainst the viral spike protein."Our studyand the new clinical trial data show thatthe virus is traveling in a direction thatis causing it to escape from our currentvaccines and therapies that are directedagainst the viral spike, said Ho. "lf the rampant spread of the viruscontinues and more critical mutationsaccumulate, then we may be condemnedto chasing after the evolving SARS-CoV-2continually, as we have long done forinfluenza virus," Ho explained."We have tostop the virus from replicating and thatmeans rolling out vaccine faster and stickingto our mitigation measures like masking andphysical distancing. Stopping the spreadof the virus will stop the development offurther mutations" Synthetic Nanobodies,"Sybodies,Found That Neutralize SARS-CoV-2 nterrupting the interaction between thespike protein on the SARS-CoV-2 virus andits receptor on host cells, ACE2, has becomethe key target of COVID-19 therapeutics.Therapeutic neutralizing antibodies achieve this,constituting a potential treatment. However,antibody production is traditionally a somewhatslow and expensive process.Now,a Germangroup of researchers reports the rapid isolationand characterization of nanobodies from a synthetic library, known as sybodies,that targetthe receptor-binding domain (RBD) of theSARS-CoV-2 Spike protein. This work is published in NatureCommunications in a paper titled,"Selection,biophysical, and structural analysis of syntheticnanobodies that effectively neutralizeSARS-CoV-2" To enable the virus to hook onto the cellsurface, the spike protein binds ACE2 using three finger-like protrusions, called thereceptor-binding domains (RBDs). Blocking theRBDs therefore has the potential to stop thevirus from entering human cells.This can bedone using antibodies. Nanobodies, small antibodies found in camelsand llamas, are promising as tools againstviruses due to their high stability and small size.Although obtaining them from animals is timeconsuming, technological advances now allowfor rapid selection of synthetic nanobodies,called sybodies. A technology platform to selectsybodies from large synthetic libraries wasrecently developed in the lab of Markus Seeger,PhD,assistant professor at the University ofZurich, and made available for this study. In search of the best sybodyagainst SARS-CoV-2 The lab of Christian Low, PhD, group leader atEMBL Hamburg, searched through the existinglibraries to find sybodies that could blockSARS-CoV-2 from infecting human cells. First,they used the viral spike protein's RBDs as bait toselect those sybodies that bind to them. Next,they tested the selected sybodies according totheir stability, effectiveness, and the precisionof binding. Among the best binders, one calledsybody 23 turned out to be particularly effectivein blocking the RBDs. "Several binders with low nanomolar affinities Nanobodies, small antibodies found in camels and llamas, arepromising as tools against viruses but time consuming to obtainfrom animals. and efficient neutralization activity wereidentified of which Sb23 displayed high affinityand neutralized pseudovirus with an IC50 of0.6 ug/mL"the authors wrote. To learn exactly how sybody 23 interacts withthe viral RBDs, researchers in the group of DmitriSvergun, PhD, senior scientist at EMBL Hamburg,analyzed the binding of sybody 23 to the RBDs bysmall-angle X-ray scattering. In addition, MartinHallberg, PhD, at the Karolinska Institutet usedcryo-EM to determine the structure of the fullSARS-CoV-2 Spike bound to sybody 23. The authors explained that the cryo-EMstructure of the spike bound to sybody 23showed that"sybody 23 binds competitively inthe ACE2 binding site" Furthermore,they noted, "the cryo-EM reconstruction revealed an unusualconformation of the spike where two RBDs are inthe'up'ACE2-binding conformation." The RBDs switch between two positions: in the"up"position the RBDs poke out, ready to bindACE2; in the"down"position they are furled to hidefrom the human immune system. The molecularstructures revealed that sybody 23 binds RBDsin both"up"and"down"positions,and blocks theareas where ACE2 would normally bind. This abilityto block RBDs regardless of their position mightexplain why sybody 23 is so effective. Finally, to test if sybody 23 can neutralizea virus, the group of Ben Murrell at KarolinskaInstitutet used a different virus, called a lentivirus,modified such that it carried SARS-CoV-2's spikeprotein on its surface. They observed that sybody23 successfully disabled the modified virus invitro.Additional tests will be necessary to confirmwhether this sybody could stop SARS-CoV-2infection in the human body. Scientific collaborationduring lockdown "The collaborative spirit has been enormousin these times, and everybody was motivated tocontribute,said Low. The researchers started theproject as soon as they received approval fromEMBL leadership to reopen their laboratoriesduring the COVID-19 lockdown. They managedto select the candidate sybodies and perform theanalyses in just a few weeks. “Getting the results so quickly was onlypossible because the methodologies we usedhad already been established for other researchprojects unrelated to SARS-CoV-2. Developingthese tools would have taken significantly moretime and resources"said Low. The results of this project hold out the promiseof a potential way to treat COVID-19. In futurework, the scientists will perform further analysesto confirm whether sybody 23 could be aneffective COVID-19 treatment. Abstract Since COVID-19 was first discovered in late 2019, millions of people have been infected, and hundredsof thousands of deaths reported so far. Controlling the outbreak and disease progression in infectedpersons requires the development of vaccines and therapeutics in accelerated timelines. In this article,we review a few selected studies on SARS CoV 2 structural biology, host receptor recognition properties,cross-reactivity with SARS CoV and MERS CoV neutralization antibodies, and several workflows aimed atdeveloping neutralization antibodies and therapeutics and how Octet@ label-free platforms are utilized toaccelerate experimental timelines. OctetBio-LayerInterferometrySystems: AdvancingDevelopment ofCoronavirus Vaccine and Therapeutics Introduction The World Health Organization declared thenew COVID-19 outbreak as a pandemic onthe 11th of March 2020. Millions of people havebeen infected worldwide with the virus, andhundreds of thousands of deaths reported sofar. The rapidly evolving epidemiology of thepandemic and absence of a licensed therapeuticor vaccine for the disease has accelerated theneed to develop an intervention. At the heart ofthe search for a vaccine are modern biochemicaland analytical technologies that can be deployedfor the rapid screening of the many potentialcandidates to determine those with the best chances of success. Since the COVID-19 genomesequence was first shared in January 2020,Octet" system data has been featured in severalbreakthrough publications related to coronavirusbiology, vaccine and antiviral therapeuticdevelopment. Octet"BLI platforms are helping accelerateSARS CoV-2 research by providing scientists: High-throughput kinetic interaction analysissolutions to select and characterize antigenictargets, vaccine and therapeutic candidates fast Fast and flexible platform for antibodyepitope characterization and coveragedetermination Efficient workflows with a wide rangeof biosensors tailored for vaccine andbiotherapeutic development research Versatile solutions in upstream anddownstream vaccine development andmanufacturing such as vaccine potency,stability and titer measurements Intuitive, easy-to-use data analysissoftware tools Octet@ instruments utilize Bio-Layer Interferometry (BLI) technology that measureslight interference as a result of binding ona sensing surface (biosensors) to monitorbiomolecular interactions.The detection is reported in real-time as binding responses.The instrument operates in a microfluidics-freeenvironment where samples are housed in 96-or384-well plates. Assay workflow involves a simpleDip and Read measure, where the disposablebiosensor is dipped into samples presented inmicrowell plates, and interactions are detected inreal-time. This workflow is advantageous overmicrofluidics based technologies in that itenables specific capture and analysis of targetmolecules in complex matrices such as cellculture supernatants, serum, plasma andother media, circumventing the need to purify antibodies and other recombinant proteins forscreening without the risk of sample clogging.These features enable users to easily vary assayand biosensor conditions and reuse precioussamples that result in fast and low-cost assayoptimization. Additionally, users can choose from a rangeof tailored affinity capture and immobilizationchemistries suitable for vaccine andbiotherapeutic development research.Widelyused,ready-to-use biosensor chemistries arelisted in Table1. Table 1: Widely used biosensor chemistries forantibody-based kinetic analysis, receptor binding and epitope binning assays. See full selection ofbiosensor chemistries. Octet@ instruments have been widelydeployed in various vaccine developmentresearch programs (HIV1,2, influenza, Ebola4,novel coronavirus5-15) and in evaluating vaccineagents such as nanoparticles16, virus-likeparticles (VLPs)17 and bispecific antibodies18.Octetsystems also provide solutions beyondlead candidate selection to bioprocessdevelopment and manufacturing. This reviewsummarizes the use of Octetassays in somerecent advances in coronavirus research andvaccine development efforts. Table 1:Widely used biosensor chemistries for antibody-based kinetic analysis, receptor binding and epitope binning assays. See fullselection of biosensor chemistries. Biosensor Description Application AHC Anti-Human Fc-Capture Capturing human IgGs or human Fc-fusion proteins from purified or unpurified media for kinetic or epitope binning analysis with various analytes. AMC Anti-Mouse Fc-Capture Capturing mouse lgGs or mouse Fc-fusion proteins from purified or unpurified media for kinetic or epitope binning analysis with various analytes. SA Streptavidin Immobilizing biotinylated molecules for allkinetic or epitope binning analysis. HIS1K Anti-Penta-HIS Capture of His-tagged proteins from purified or unpurified media for kinetic ana- lysis with target analytes. Quantitation of His-tagged proteins in buffer, media or diluted lysate. Biosensor is pre-coated with Penta-His antibody. FAB2G Anti-Human Fab-CH1 Kinetic analysis of human Fab fragments and IgG with target antigen, Fc receptors, 2nd Generation or other analytes. Quantitation of Fab and lgG. Quantitation of HIS-tagged proteins in buffer or diluted matrix, capturing of HIS- tagged proteins for kinetic analyses with various analytes. NTA Ni-NTA Protein A Fc-Capture Capturing lgGs and Fc fusion proteins from various species including human. Determination of CoronavirusReceptor Binding Mechanismsand Cross-Reactivity The 2019-nCoVor SARS CoV-2 Shares geneticand morphologic features with other coronavirusfamilies, particularly fromthe betacoronavirusgenus (SARS-CoV 2003). SARS CoV-2 makes use ofa glycosylated, homotrimeric class l fusion spike(S) protein to gain entry into host cells via thehuman angiotensin converting enzyme-2 (ACE2)receptor19. The SARS CoV-2 S glyco-protein bearssignificant structural homology with SARS-CoVcompared to other major coronaviruses such asthe MERS-CoV. The S-protein is known to exist in a metastablepre-fusion conformation that undergoesrearrangements to fuse the viral membrane withthe host cell membrane. Binding of the S1 subunitto host-cell receptors triggers destabilization ofthe pre-fusion trimer and conformational changein the protein initiates fusion with the host cellmembrane through the S2 subunit. In order toengage the host-cell receptor,the receptor-binding domain (RBD) of S1 would need toundergo hinge-like transitions that either hidesor displays the receptor-binding regions whichare termed as down and up conformationsrespectively, providing accessibility to receptor-binding and presenting itself as a key antigenictarget for vaccine development. One of the first CryoEM structures of the SARS CoV-2 S trimer in the pre-fusion conformationwas revealed by Wrapp et al. where they showedthat whilethe overallstructure of SARS CoV-2 Sprotein was mostly similar to SARS-CoV S,differences existed between the downconformations at the RBD1. Another cryo-EM structure of SARS CoV-2 Spikeglycoprotein complex resolved by Walls et al.showed that that the SARS-CoV-2 S glycoproteinharbors a furin cleavage site at the boundarybetween the S1/S2 subunits, which is processedduring biogenesis and differentiates this virus apartfrom SARS-CoV and other SARS-related CoVs10Using Octet binding assays, they showed thatthe receptor-binding domains of SARS-CoV-2 Sand SARS-CoV S engaged the human ACE2 withsimilar binding affinities. Similar observations werereported by Joyce et al. using a high-resolutioncrystal structure of the SARS-CoV-2 S RBD.Additionally, in this work antibodies that wereknown to interact with SARS-CoV and MERS CoVreceptor binding domains (RBD) were testedfor SARS-CoV-2 binding activity using an Octet@kinetic assay.Only two antibodies, 240CD andCR3022 out of the pool, displayed low nanomolarbinding affinities against the SARS-CoV 2 RBD. These two antibodies, however, competedfor the same SARS-CoV-2 RBD binding siteaccording to an Octet°cross-competition assay.Cross-competition or epitope binning assayscan be used to segment monoclonal antibodies A B In-tandem assay Classical sandwich Premix assay assay Classical Sandwich In-tandem (mAbs) into bins based upon the antigen region or epitope bound by each antibody (Figure 1).Furthermore, an Octet@competition assay revealedthat 240CD or CR3022 antibody binding to SARSCoV-2 RBD did not perturb ACE2 recognition. Crystallography data revealed that CR3022engaged the RBD at a site that is conservedin both SARS CoV and SARS CoV-2 explainingthe cross-reactivity. However, a comparison ofSARS-CoV and MERS-CoV epitope regions fromprevious structural data to SARS CoV-2 identifiedin this work revealed that CR3022 was binding toa novel epitope within SARS CoV-2 S. A specific RBD knockout mutant introducedwithin this epitope region (glycan sequon atposition 384) eliminated binding to both CR3022and 240CD antibodies according to Octetdata confirming shared epitopes between bothantibodies. To understand the SARS CoV-2 Spikeconformations with respect to receptor bindingand cell entry, structures of CR3022 bindingto trimeric units of SARS-CoV-2,SARS-CoV andMERS-CoV were modeled5. Data indicated thatCR3022 epitopes were obstructed by adjacent spike promoters when the RBD is in the downconformation but were more accessible in theup confirmation.Octetassays observed CR3022binding to non-stabilized S-glycoprotein (S1)conformations and much weaker binding to S2P trimers. To assess whether minimal proteolyticaction or receptor binding could increase theavailability of the obscure CR3022 epitope, S2trimers were treated with trypsin or incubatedwith ACE2. Incubation of S2 P trimer withhuman ACE2 did not dramatically affect CR3022binding. However, trypsin treatment of the S2P conformation resulted in increased bindinglevels similar to non-stabilized S glycoproteinbinding, and the level of binding was titratablewith increasing proteolytic action, inferring to thecryptic nature of the CR3022 binding epitope.Similar observations were reported by Yuan et al.,using the crystal structure of neutralizing antibodyCR3022 bound to SARS-CoV-2 RBD14. Octet@ binding data showed that despite highsequence conservation between SARS-CoV andSARS CoV 2 RBDs, CR3022 FABs binds SARS-CoVwith ~100-fold higher affinity than to SARSCoV-2 RBD possibly driven by the non-conserved Figure 1:Assay formats of epitope binning assays and recommendation guide to select the most suitable epitope binning assay format.Octetsystems enable multiple assay formats for cross-competition assessment and provide dedicated high-throughput software analysistools for epitope binning. (A) In Tandem Assay: Antigen is immobilized on the biosensor, followed by the binding of the saturating mAb(Ab1) and competing mAb (Ab2), respectively. (B) Classical Sandwich Assay: One mAb is immobilized on the biosensor (Ab1), antigen iscaptured using this mAb and then a second sandwiching mAb is tested (Ab2). (C)Premix Assay: One mAb is immobilized on the biosensor(Ab1), the biosensor is then exposed to a premix solution containing the antigen and a large molar excess of the second mAb (Ab2). (D)Guidance for selecting the most suitable epitope binning assay format. Please read the Octetassay guide to learn more about developingand analyzing cross-competition screens. residues between the two epitopes mentionedabove. An in vitro microneutralization assayindicated higher neutralization activity withSARS-CoV, but not SARS-CoV-2 for CR3022, aresult that is consistent with lower RBD bindingaffinities.Overall, these structural studies provideinsight into how SARS CoV-2 can be targeted bythe humoral immune response and revealed aconserved, but cryptic epitope shared betweenSARS CoV-2 and SARS CoV that can potentially beutilized to create cross-protective nAbs. Neutralization Antibody Development Neutralizing antibodies (nAbs) are likely to beone of the most effective treatments againstcoronavirus infections among the currenttherapeutic options and are in urgent need.Several nAbs targeting SARS-CoV RBD, have beenshown to exhibit significant in vivo antiviral activityby reducing virus titers in animal models. However,the availability of nAbs with cross-reactivity couldbe key for sustained utilization as therapeutics thatcan stand against virus mutations that are typicalof single standard RNA viruses. Due to the ~80% sequence similaritybetween the two Spike (S) proteins of SARSCoV and SARS CoV-2,understanding whetherantibodies raised from SARS-CoV spike (S) proteinimmunization would retain cross-reactivity tothe new SARS-CoV-2 will offer important insightsand guidance to therapeutic antibody and prophylactic vaccine development. Tian et al., in a pioneering study, expressedand purified the CoV-2 RBD and tested thebinding of several SARS-CoV specific neutralizingantibodies that were known to target the RBDand exert potent neutralization activities againstSARS-CoV9. SARS-CoVnAbs,m396,CR3014,CR3022 and the MERS-CoV nAb m336 weretested for SARS-CoV-2 RBD recognition (Figure 2).While most antibodies did not show appreciablebinding to the SARS CoV-2 RBD, CR3022 that wasisolated from a SARS CoV patient bound with a6.3 nM affinity(k of1.84×10Ms-and kof1.16×10-3s-1). Octet@ BLI competition assays showed thatCR3022 did not compete for the ACE2 bindingsite in SARS-CoV-2 RBD, indicating binding toan epitope different from the ACE2 binding site(Figure2B). Similarly, Wrapp et al. investigatedanother set of previously published SARS-CoVRBD binding antibodies, S230, m396 and 80R withSARS CoV 2 S protein12.Octet@ binding assaysindicated a lack of significant binding activitypossibly due to the recognition through differentepitopes compared to SARS CoV. Sun et al.reported that several pAbs and mAbs generatedby immunizing mouse and rabbits with SARS-CoVS1 or RBD proteins did not show favorablecross-neutralization activities in SARS-CoV-2pseudoviruses (PSV). However, these antibodiesexhibited potent neutralization potencies against Figure 2: Characterizing of SARS CoV antibody interactions to SARS CoV-2 RBD. (A) Binding profiles of SARS CoV-2 RBD to ACE2 andantibodies and (B) competition of CR3022 and ACE2 with SARS CoV-2 RBD measured by the Octet° system. For the competition analysis,ACE2 were immobilized onto HIS1K biosensors, followed by binding to either ACE2, CR3022 + ACE2 or isotype antibody control of ACE2.Figure is reproduced from reference 9. SARS CoV virus and high-affinity binding toSARS CoV RBD (K~9-200 pM)8. These resultsindicate that antibodies that exhibit SARS-CoVneutralization potencies or RBD binding is notnecessarily a prerequisite for activity in SARS CoV-2despite high sequence homologies at the RBD andsuggests the necessity to develop novel antibodiesthat specifically target SARS CoV-2 RBDs. Wang et al. reported the discovery of ahuman monoclonal antibody with neutralization potencies for SARS-CoV-2 and SARS-CoV virusactivity.47D11, derived from a screen thatconsisted of a collection of supernatants thatcontained SARS S hybridoma derived fromimmunized transgenic mice, showed cross-reactivity and neutralization activity against bothSARS CoV and SARS CoV-2 pseudotypedVSVinfections.Octet@ instrument studies showedthat 47D11 binds the SARS CoV S domain withhigher affinity (K = 0.745 nM) relative to SARS CoV-2 S domain (K =10.8 nM) although similarbinding affnities were reported for bindingto the SARS CoV and SARS CoV-2 RBD bindingdomains, further validating the differences inepitope accessibility between the two virusreceptor binding motif (RBM) conformations.Additionally, as in other cases reported thus far,47D11 did not compete with the SARS CoV orSARS CoV-2 RBD, implying neutralization througha mode that is different from receptor blocking.This cross-neutralizing antibody reportedlytargets a communal epitope and offers potentialfor prevention and treatment of SARS CoV-2. Pinto et al. screened for nAbs from a SARS CoVsurvivor . Eight mAbs from the screen boundto both SARS CoV and SARS CoV-2 S protein-transfected CHO cells, out of which mAbs S303,S304, S309 and S315 recognized both SARSCoV and SARS CoV-2 RBDs.Octet system dataindicated S309 binding to both S domains withsimilar nanomolar affinities and also showedcomparable neutralization potencies againstboth SARS CoV and CoV-2 pseudo-viruses andauthentic SARS-CoV-2. According to structuraldata, S309 recognizes a protein/glycan epitopeon the SARS CoV-2 RBD distinct from thereceptor-binding motif (RBM) and seems to beaccessible to both up and down states. Octet@ BLI competition assays showed thatbinding of S309 Fab and IgG forms to SARS CoV-2RBD did not discourage ACE2 receptor binding to SARS CoV-2 S protein. To gain more insight onthe epitopes recognized by the panel of mAbs, anOctet epitope binning assay was carried out tomap the antigenic sites present on the SARS CoVand,SARS-CoV-2RBDs. Octet epitope binning analysis identifiedfour antigenic regions within the RBD ofSARS CoV where the mAbs were targeting.Based on these findings, mAb combinationsthat targeted different antigenic regions weretested for neutralization potencies and possiblesynergistic effects where data indicated thateither S304 or S315 in combination with S309enhanced neutralization potencies againstSARS CoV-2. S309 identified in this work showedbroad neutralization activity against multiplesarbecoviruses showing promise as an effectivetherapeutic candidate. Pan et al. used the SARS CoV-2 RBD domainto produce neutralization antibodies in horseantisera.F(ab),s isolated from horse antiserareportedly showed neutralization activity againstSARS CoV-2 with an ECg of ~25 ug/mL.OctetBLI kinetic analysis also confirmed SARS CoV-2receptor binding to F(ab), with an affinity of 76nM. Affinity purification of F(ab'), from antiseraimproved neutralization activity against SARSCoV-2 (EC80=0.18 ug/mL) and increasedbinding affinity (0.76 nM) measured by Octet BLI,highlighting RBD-specific F(ab), as a potentialtherapeutic candidates for SARS CoV-2. Wu et al. reported the generation and testing ofsingle-domain antibodies targeting SARS CoV-2RBD13.The panning using SARS-CoV-2 RBD andS1 as antigens resulted in the identification ofantibodies targeting five types of neutralizing ornon-neutralizing epitopes on SARS-CoV-2 RBD.They tested 18 human single-domain antibodiesin competition binding assays using the Octet@system and found that they could be divided intothree competition groups (groups A, B, or C) thatdid not show any competition with each other.The single domain antibody n3130 identified fromthe library using SARS-CoV-2 S1 as the panningantigen showed potent neutralization activitywith both pseudotyped and live virus and strongbinding to SARS CoV-2 S1 and RBD domains.This work also reports the unique immunogenicprofiles of SARS-CoV-2 RBD compared to thatof SARS-CoV and MERS-CoV, which may haveimportant implications for the development ofeffective vaccines against SARS CoV-2. Design and Validation of a PeptideInhibitor to Block Viral Entry In addition to protective antibodies, peptideand small molecule inhibitors can also be usedas viable inhibitory strategies to target the SARSCoV-2 RBD to prevent virus entry. Zhang et al.investigated the design and development of a23-mer peptide inhibitor (SBP1) discovered basedon the ACE2 PD a1 helix that spans the binding interface according to the crystallographicstructure of the SARS-CoV-2 S receptor-ACE2protein complex15. Octet BLI was utilized to the test targetspecificity of the peptide binding to SARS CoV-2RBD.SPB1 bound the RBD with an affinity of 47nM (k=4.69×104M-sand k=2.2×10s) where this binding affinity is comparableto that of the full-length ACE2 binding to SARSCoV-2 RBD. SBP1 could, therefore, potentiallycover spike proteins on the SARS-CoV-2 Surfaceand outcompete the binding for ACE2 as a noveltherapeutic intervention strategy. Summary Affinity characterization, neutralization andcompetition studies are essential in the selectionof high affinity and potentially neutralizinglead candidates for therapeutic and vaccinedevelopment against the novel SARS CoV-2.Kinetic analysis further describes the componentsof association and dissociation that comprise theoverall affinity, while cross-competition assaysallow for the identification of more optimalcandidates. In addition to the studies illustrated above.researchers have also utilized Octet@ assays tocharacterize and investigate other members ofthe coronavirus or associated species where theobservations and results are invaluable towardsimplementing and framing current therapeutic Table 2: Examples of Octet° BLI assays in coronavirus research and vaccine development. strategies for SARS CoV-2. Table 2 providesa summary of selected studies illustratingtheuse of Octet@ assays on current and historicalcoronavirus research20-27. In all these studies, theOctetsystem's ease-of-use combined with the References ( 1. S tructures of protective antibodies rev e al sit e s of vulnerability on E bola virus, Murin C D et al.,Proc Natl Acad S ci USA, 111:17 1 82-17187,2014, doi:10.1073/pnas.1414164111. ) ( 2. A n ext- g eneration cleaved, soluble H I V-1 E nv trimer, BG505 S OSIP.664 gp140, expresses multiple epitopes for broadly neutralizing butnot non-neutralizing antibodies, Sanders RW, et al.,PLoS Pathog, 9:e1003618,2 0 13, doi:1 0 .1 3 71/journal.ppat.1 0 03618. ) ( Kineti c analysis of the inf l uenza A virus HA/NA balance reveals contribution of NA to virus-re c eptor binding and NA-depende n trol l ing on r eceptor-co n taining surfaces, Guo H, et al . , PLo S Pathog, 14:e1007233,2018, doi:10.1371/journal.ppat.1007233. ) ( 4 Antibody Repertoires to the Same Ebola Vaccine Antigen Are Differentially Affected by Vaccine Vectors, Me y er M, et al., Cell Rep, 24: 1 816-1829,2018, doi:10 . 1016/j.celrep. 2018.07.044. ) ( 5 A Cryptic Site of Vulnerability o n the Receptor Binding Domainof t he S ARS-C o V-2 Sp i ke Glycoprotein, J o yce MG, et al., bioRxiv, 2020.2003.201 5 .992883,2020, doi:10.1 1 01/2020.03.15.992883. ) ( .6 Immunoglobulin fragment F(ab')2 against RBD potently neutralizes S ARS-CoV-2 in vitro, Pan X, e t al., bioRxiv, 2020 . 2004.2007.029884,2020, doi:10.1101/2020.04.07.029884. ) 7. Structural and functional analysis of a potent sarbecovirusneutralizing antibody, Pinto D, et al., bioRxiv, 2020.2004.2007.023903,2020, doi:10.1101/2020.04.07.023903. 8. SARS-CoV-2 and SARS-CoV spike-RBD structure and receptor bindingcomparison and potential implications on neutralizing antibody andvaccine development, Sun C, et al., bioRxiv, 2020.2002.2016.951723,2020,doi:10.1101/2020.02.16.951723. ( 9. Potent bindi n g of 2019 novel coronavirus spike protein by a SARScoronavirus-specific human monoclonal antibody, Tian X, et a l., Emerg Microbes I nfect, 9:38 2 -385,2020,doi:10.1080/2222 1 751.2020. 1 7290 69. ) ( 10. Structure, function, and antigenicit y of the SARS-CoV-2 Spike gl y coprotein, Walls, A C e t al., C ell,181:281-292 e286,2020, doi:10.1016/j.c e ll.2020.02.058. ) 11. A human monoclonal antibody blocking SARS-CoV-2infection,Wang C, et al., bioRxiv, 2020.2003.2011.987958, 2020,doi:10.1101/2020.03.11.987958. 12. Cryo-EM structure of the 2019-nCoV spike in the prefusionconformation, Wrapp D,et al., Science, 367:1260-1263,2020,doi:10.1126/science.abb2507. 13. Fully human single-domain antibodies against SARS-CoV-2,Wu Y, et al., bioRxiv, 2020.2003.2030.015990, 2020,doi:10.1101/2020.03.30.015990. availability of a variety of ready-to-use biosensorchemistries has enabled users to significantlyreduce time to results and advance thecontinued search for vaccines and therapeuticsfor the current pandemic. ( 1 4 . A highly conserved cryptic epitope in the receptor-b i nding domains o f SARS-CoV-2 and SARS-CoV, Science,doi:10.1126/science.abb7269 (2020). ) ( 15. The first - in - class peptide binder to the SARS-C o V-2 Spike p rotein, Zhang G, et al., bioRxiv, 2020.2003.2019.999318,2020, doi:10. 1 101/2020.03.19.999318. ) ( 16. P resenting n a tive-li k e trimeric HIV-1 antigens with self-assembling nanoparticle s , He L, et al., Nat C ommun, 7: 1 2041, 2016, doi:10.1038/ ncomms12041. ) ( 1 7 . Exposure of epitope residues on the outer face of the chikungu n yavirus envelope trimer determines antibody neutralizing efficacy, Fong R H, e t a l., JVirol, 88:14364 - 14379,2014, doi:10.112 8/ JVI.01943-14. ) ( 18. A "Tr o jan h o rse"bispecific-antibody strategy for broad protection against e bolaviru s es, Wec AZ, et al., Science, 354:350-354,2016,doi:10.1126/science.aaq3267. ) ( 19. F unctional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses,Letko M, Marzi Aand Munster V, Nat Microbiol, 5:562- 5 69,2020, d o i:10 . 10 3 8/s41564- 020 -0 688-y. ) 20. Structural basis for human coronavirus attachment to sialic acidreceptors, Tortorici MA, et al.,Nat Struct Mol Biol, 26:481-489,2019,doi:10.1038/s41594-019-0233-y. 21. Potent MERS-CoV fusion inhibitory peptides identified from HR2domain in spike protein of bat coronavirus HKU4, Xia S, et al.,Viruses,11,2019, doi:10.3390/v11010056. 22. Towards a solution to MERS: protective human monoclonalantibodies targeting different domains and functions of the MERS-coronavirus spike glycoprotein, Widjaja l, et al., Emerg Microbes Infect,8:516-530,2019,doi:10.1080/22221751.2019.1597644. ( 23. I mportance of neutralizing m onoclonal antibodies targeting multip l eantigeni c sites on the Middle East Respiratory Syndrome coronavirusspike glycoprotein to avoid neutralization escape, Wang L, et al., J .Virol, 92, 2018, doi:10 . 1128/JVI.02002-17. ) 24. Structural definition of a neutralization epitope on the N-terminaldomain of MERS-CoV spike glycoprotein, Zhou H, et al., Nat Commun,10:3068,2019, doi:10.1038/s41467-019-10897-4. 25.U1ltrapotent human neutralizing antibody repertoires against MiddleEast Respiratory Syndrome coronavirus from a recovered patient, NiuP,et al., J Infect Dis, 218:1249-1260,2018, doi:10.1093/infdis/jiy311. ( 26. E valuation of candidate vaccine approaches for M ERS-CoV, Wang L, et al., Nat Commun,6:7712,2015, doi:10.1038/ncomms8712. ) 27. A humanized neutralizing antibody against MERS-CoV targetingthe receptor-binding domain of the spike protein, LiY, et al, Cell Res,25:1237-1249,2015, doi:10.1038/cr.2015.113 Accelerate Your Antibody Developmentand Still Have Peace of Mind Gain More Insights, Make Better Decisions.Sartorius accelerates development of breakthrough therapeutics withgroundbreaking, comprehensive solutions for lab filtration, cell/proteinanalysis, and more. Our products and technologies provide rapid,multifactorial results to enable deeper insights and ensure the choice,quality, and integrity of your biopharmaceutical product. Advance time to results with: Process streamlinina "Multiplexing mInformation-rich data Biologicallyrelevant insights www.sartorius.com/mab-development Simplifying Progress SARTORIUS |GENengnews.comnI 牛IgG具有高达70个氨基酸的超长CDR-H3结构域。之前发现,具有牛的CDR-H3的HIV中和抗体会识别gp120 CD4结合位点上的表位,该表位通常凹陷于常规抗体。所以超长CDR-H3抗体可能会识别一些人抗体无法识别的隐蔽表位,并扩大抗体识别的靶点区域[1]。应用案例德国默克(Merck)的科学家们设计了基于牛CDR-H3双特异性抗体,靶向肿瘤细胞上的表皮生长因子受体(EGFR)以及自然杀伤(NK)细胞上的天然细胞毒性受体NKp30。基于牛IgG的双特异性抗体(BsAbs)引发了有效的NK细胞重定向,从而引发了EGFR过表达的肿瘤细胞裂解以及促炎细胞因子的强劲释放[2]。牛源CDR-H3双特异性抗体的产生示意图。通过牛抗体文库筛选出针对两个靶标(以红色和黄色显示)的识别序列。嫁接到人源Fab支架上,利用重链异源二聚技术(例如SEED技术)组装成双特异性抗体。主要技术路线Octet®高通量筛选高亲和力双特异性抗体Octet®基于生物层干涉技术(BLI),是被美国药典认可的用于亲和力检测的标准方法。Octet®具有高通量和易使用等特点,成为生物医药企业必备工具之一。在Merck的这个方案中,Octet®被选择作为抗体亲和力检测的主要方法。Octet® Red 96,用Anti-human Fc传感器固化抗体,分别与EGFR(左图),NKp30(中图)进行结合解离检测其亲和力。然后通过连续结合两个靶点,观察抗体是否可以同时结合两个抗原(右图)。用Octet®检测的亲和力结果列表(部分)iQue®高通量流式检测细胞水平结合科研人员使用iQue® 3高通量流式细胞仪评估细胞结合,并使用仪器配套智能软件ForeCyt®进行分析。iQue®高通量流式具有检测速度快,样品耗量少等特点,是细胞水平高通量筛选的常用仪器。左图为EGFR高表达的A431细胞与双特异抗体结合:每孔测量800-1800个A431细胞。在两个初始洗涤步骤后,将细胞和100 nM的bsAbs孵育,洗涤后加入荧光标记的Goat Anti-Human IgG和碘化丙啶(标记死细胞)。可见,bsAbs可以和A431细胞结合。右图为双特异抗体与NKp30结合:检测双特异性抗体是否可以同时结合两个靶点,在bsAb以100 nM的浓度与A431细胞孵育和洗涤后,与His标签的NKp30孵育30分钟。再将细胞与抗His抗体和碘化丙啶孵育。可见,bsAbs可以同时结合A431细胞和NKp30。Incucyte®实时监测杀伤效果对于免疫细胞杀伤实验,Incucyte®实时活细胞分析系统可以在不移动细胞培养板的情况下,对细胞状态进行实时荧光拍摄,不需要对细胞进行悬浮处理,并且具备高通量特性,可同时放置6块96或384孔板。A431细胞深红色染料染色。将靶细胞以20 μL体积以2500个细胞/孔接种到384孔透明底部微量滴定板中,并孵育3小时。以不同的E:T比(即1:1,5:1,10:1和20:1)加入NK细胞,再加入不同浓度的BsAbs。用绿色凋亡染料进行死活细胞染色,然后在Incucyte®系统中进行实时在线测量24小时。计算红绿叠加面积为裂解率。用西妥昔单抗触发的最大裂解率或用30 μM星孢菌素培养的靶细胞进行normalization。可见,活性最佳的几个BsAbs显示出nM级的EC50杀伤效果(A图),接近于100%的最大杀伤率(B,C图)赛多利斯生物分析部门始终致力于智能、可靠的高通量药物表征及筛选方法的开发。无论在药物研发还是在基础研究中,生物分析三剑客—Octet®分子互作分析系统、iQue®高通量流式细胞仪和Incucyte®实时活细胞分析系统,都能够分别从分子互作水平、细胞互作水平和细胞成像水平中提供更高效,更便捷的解决方案,让大家在竞争激烈的市场中占有一席之地!Download了解更多生物分析三剑客在抗体开发与表征中的应用,下载电子书《Neutralizing Antibodies Making Their Mark in Next Wave of Biologics》点击下载 获取资料-参考文献-Structural Basis of Broad HIV Neutralization by a Vaccine-Induced Cow Antibody. Sci Adv (2020) 6:eaba0468. doi: 10.1126/sciadv.aba0468Grabbing the Bull by Both Horns: Bovine Ultralong CDR-H3 Paratopes Enable Engineering of 'Almost Natural' Common Light Chain Bispecific Antibodies Suitable For Effector Cell Redirection. Frontiers in Immunology, 01 Jan 2021, 12:801368
确定

































还剩40页未读,是否继续阅读?
德国赛多利斯集团为您提供《抗体中开发与表征检测方案(大分子作用仪)》,该方案主要用于其他中表征检测,参考标准--,《抗体中开发与表征检测方案(大分子作用仪)》用到的仪器有赛多利斯 Octet® R2 生物分子相互作用分析系统、赛多利斯 CellCelector 全自动无损细胞分离系统、赛多利斯 Incucyte® SX5 活细胞分析仪、赛多利斯 Octet® R8 生物分子相互作用分析系统
相关方案
更多
该厂商其他方案
更多
























