方案详情
文
短短几十秒内就可彻底裂解粪便中的微生物;
得到的DNA可直接用于消化道菌群的定量和定性实验;
实验结果的可重复性及一致性;
彻底去除腐殖酸和PCR抑制剂。
方案详情

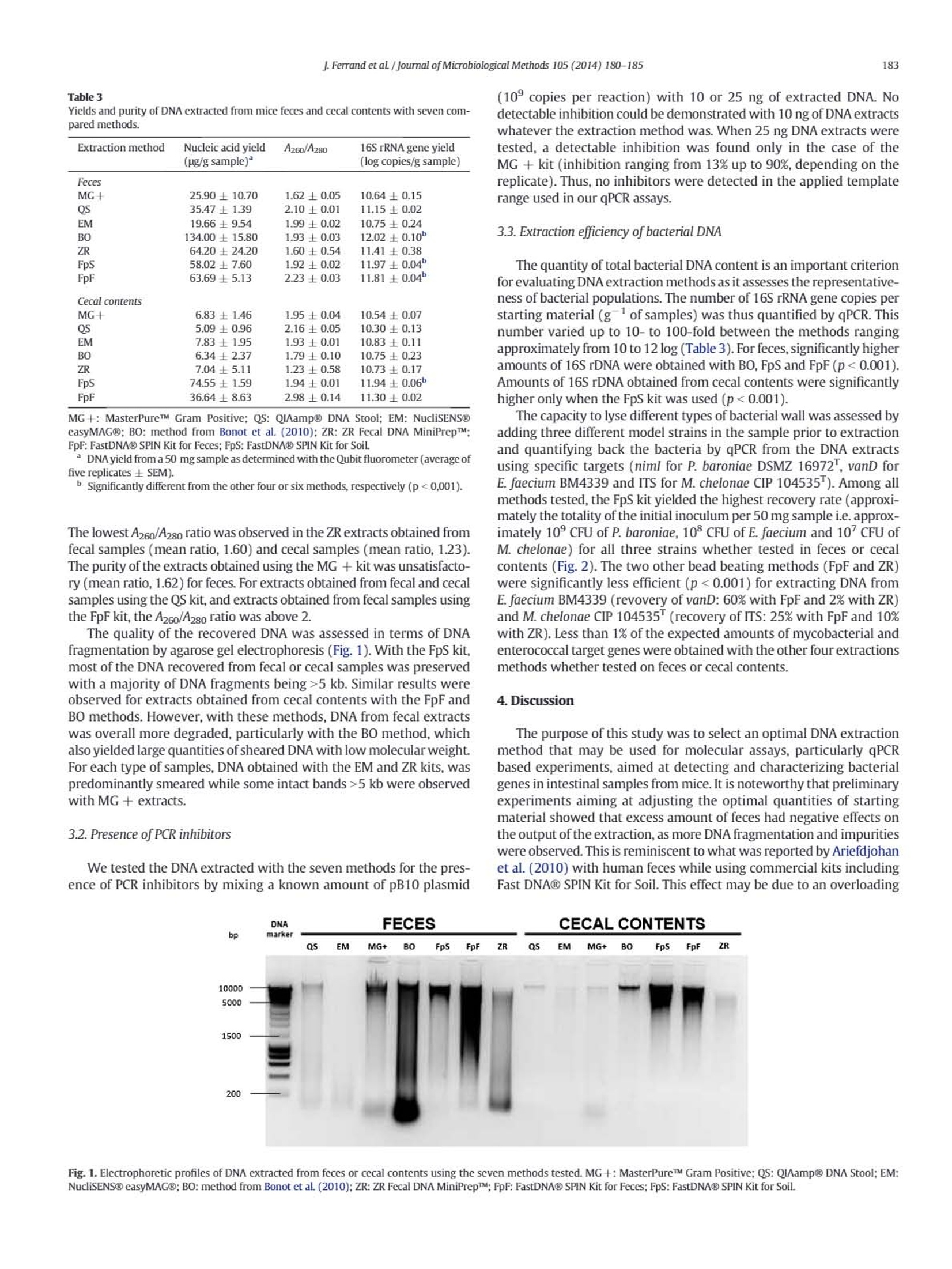
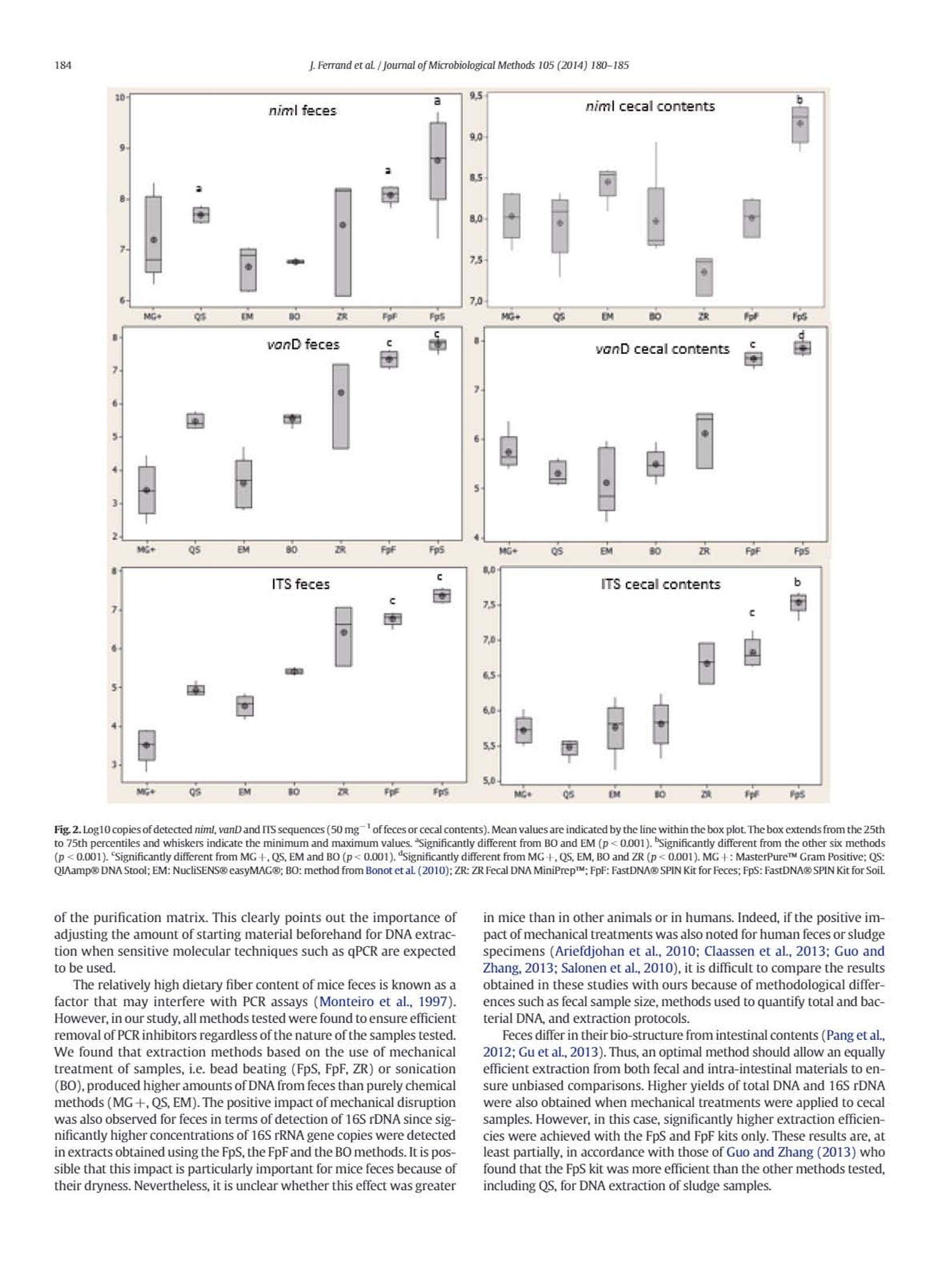
journal of Microbiological Methods 105 (2014) 180-185 181J.Ferrand et al / Journal of Microbiological Methods 105 (2014) 180-185 Contents lists available at ScienceDirect Journal of Microbiological Methods journal homepage: www.elsevier.com/locate/jmicmeth Comparison of seven methods for extraction of bacterial DNA from fecaland cecal samples of mice Janina Ferrand*, Kevin Patron, Christine Legrand-Frossi,Jean-Pol Frippiat, Christophe Merlin,Corentine Alauzet, Alain Lozniewski EA 7300 Stress Immunite Pathogenes, Universitede Lorraine, Vandoeuvre-les-Nancy, France "Universite de Lorraine-CNRS, Laboratoire de Chimie Physique et Microbiologie pour I'Environnement (LCPME), UMR, 7564 Vandceuvre-les-Nancy, France ARTICLEINFO ABSTRACT Article history:Received 7 March 2014Received in revised form 25 july 2014Accepted 25 july 2014Available online 2 August 2014 Keywords: DNA extraction Mice feces Mice cecal contents Analysis of bacterial DNA from fecal samples of mice is commonly performed in experimental studies. AlthoughDNA extraction is a critical step in various molecular approaches, the efficiency of methods that may be used forDNA extraction from mice fecal samples has never been evaluated. We compared the efficiencies of six widelyused commercial kits (MasterPureTM Gram Positive DNA Purification Kit, QIAamp@ DNA Stool Mini Kit;NucliSENS@ easyMAG@, ZR Fecal DNA MiniPrepM, FastDNA@ SPIN Kit for Feces and FastDNA@ SPIN Kit forSoil)and a non-commercial method for DNA isolation from mice feces and cecal contents. DNA quantity andquality were assessed by fluorometry, spectrophotometry,gel electrophoresis and qPCR. Cell lysis efficiencieswere evaluated by qPCR targeting three relevant bacteria in spiked specimens. For both feces and intestinalcontents, the most efficient extraction method was the FastDNA@ SPIN Kit for Soil. 16SrDNA O 2014 Elsevier B.V. All rights reserved. 1. Introduction Culture-independent molecular methods, including DNA amplifica-tion by PCR and DNA sequencing, are used in many experimental stud-ies for detecting and/or quantifying different bacterial species andgroups, as well as resistance or virulence genes in fecal samples(Alexander et al., 2011; Inglis et al, 2012; Urdahl et al., 2013). In thesestudies, obtaining sufficient amount of genomic DNA that is suitablefor further analysis is a critical factor. Both, the DNA yield and the qualityof the recovered DNA (purity and integrity) are crucial for molecular-based approaches, especially for feces that have an irregular physico-chemical structure and contain large amounts of bile salts, proteinsand complex polysaccharides that may inhibit PCR amplification.Bacte-rial DNA extraction from feces may also be hampered by incomplete celllysis because of the composition and the thickness of the bacterial cellwall. Some species can thus be underestimated or inaccessible for anal-ysis. This may particularly concern Gram-positive bacteria with thickcell wall (e.g. Firmicutes) and bacteria with mycolic acid-containingcell wall (e.g. Actinobacteria) (Maukonen et al., 2012). Mechanical dis-ruption (by bead beating, use of ultrasounds or high pressure) hasbeen reported as being the most efficient way to achieve cell wall lysis ( * C o rr espo n ding auth or a t: L a b o rat o i re d e B a ct e r i ologie, Ho p i t a l C e ntral,29, a ve n ue d u M ar echal d e L a ttre de T assigny, 5 40 00 N a nc y, Fra n ce . Tel : + 3 3 383 8 5 1 6 4 8 ; f ax: + 3 3 3 8 3 852 673. ) ( E -ma i l addr e ss: j . fer ra n d @ch u- n a n c y . f r ( J . F erra nd) . ) ( T h e s e aut ho rs contr i but e d equal l y . ) (Maukonen et al., 2012; Rapp, 2010; Salonen et al.,2010). However,this treatment may also cause DNA shearing thus limiting furthermolecular analyses (Olson and Morrow,2012). Most studies focusing on the efficiency of methods used for DNA ex-traction from feces have been performed using human (Ariefdjohanet al., 2010; Claassen et al.,2013; Nylund et al., 2010; Salonen et al.,2010; Smith et al., 2011), bovine (Rapp,2010), pig (Li et al., 2003;Tang et al., 2008) and avian (Scupham et al., 2007) samples. Mice arefrequently used in studies concerning microbialalYSIS DNA analysis of fecalsamples (Presley et al., 2010; Thompson et al.,2010;Turnbaugh et al.,2006). Surprisingly, little is known about the efficiency of good qualityDNA extraction from mice feces. Yet,several differences can be expectedas compared to other mammalian feces, notably in relation to diet type.For instance, as compared to carnivores, mice feces are particularly dry,which can impair fecal samples homogenization with lysis buffers.Moreover, rodent herbivorous diet is rich in fibers (cellulosis, hemi-cellulosis, lignin), known as PCR inhibitors (Monteiro et al, 1997),which must be efficiently eliminated during DNA extraction process. This prompted us to compare the performances of various methodsfor the extraction of DNA from mice intestinal contents and feces as, tothe best of our knowledge, this kind of study has never been performed.Six commonly used commercial methods (MasterPureTM Gram PositiveDNA Purification Kit; QIAamp@ DNA Stool Mini Kit; NucliSENS@easyMAG@; ZR Fecal DNA MiniPrepTM; FastDNAQ SPIN Kit for Feces;FastDNA@ SPIN Kit for Soil) as well as a non-commercial method,which has been successfully used for DNA extraction from complexenvironmental matrices such as sediments, activated sludge from municipal and slaughterhouse wastewater treatment plants andmanure (Bellanger et al., 2014; Bonot et al., 2010), were evaluated.The influence of these methods on total DNA yield, purity and integritywas assessed using respectively fluorometry,spectrophotometry,andgel electrophoresis. DNA extracts were also assayed by qPCR to checkfor the absence of PCR inhibitors and to determine the total 16S rDNAcopy number. Cell wall lysis efficiency was assessed by qPCR performedon DNA extracts obtained from samples that have been spiked withknown quantities of three bacterial strains differing in their cell wallthickness and composition. 2. Materials and methods 2.1. Bacterial strains Prevotella baroniae DSMZ 16972, Mycobacterium chelonae CIP104535 and Enterococcus faecium BM4339 were used for spiking offecal samples. Bacteroides thetaiotaomicron ATCC 29741 and Escherichiacoli DH5a(pB10) were used in 16S rDNA quantification and PCR inhibi-tion assays.E. coli DH5o(pB10) is a strain that contains an IncP-1Bplas-mid, pB10, that was initially isolated from sludge (Schluteret al.,2003).Prior to the assays, P. baroniae DSMZ 16972 and B. thetaiotaomicronATCC 29741 were cultured on Brucella agar (Becton Dickinson, LePont de Claix, France) under anaerobic conditions at 37 ℃ for 48 h.E. faecium BM4339 and E. coli DH5a(pB10) were cultured for 24 h onTryptic Soy Blood agar (bioMerieux, Marcy I'Etoile, France) at 37℃under aerobic conditions. M. chelonae CIP 104535 was cultured underthe same conditions but for 96 h. 2.2. Sample collection Samples were obtained from five 8-week-old C57BL/6 male mice(Charles River Laboratories, France). Feces were pooled and frozen at-20C immediately after collection until further use. Cecal sampleswere obtained shortly after dissection and immediately frozen in liquidnitrogen and stored at-80 C before use. All animal experiments wereperformed in compliance with the national and European guidelines,and approved by the Regional Ethics Committee. Samples were thawedon ice before weighing aliquots prior to further processing. 2.3. DNA extraction from mice samples With each extraction method tested, DNAs were extracted from50 mg of starting material (wet weight) in five replicates for reproduc-ibility evaluation. The characteristics of the seven methods tested aresummarized in Table 1. Among the six commercial methods, theMasterPureTM Gram Positive DNA Purification Kit (Epicenter, Madison,WI,USA) (hereafter referred as MG+), is particularly adapted toGram-positive bacteria DNA extraction. In this kit,a chemical-basedcell lysis is reinforced by the use of lysozyme. The second method testedwas the QAamp@ DNA Stool Mini Kit (Qiagen, Hilden, Germany) (hereafter referred as QS). It is based on chemical (high strengthchaotropic salts and detergents) and heat cell lysis. The NucliSENS@easyMAG@ benchtop automated nucleic acid extraction system(referred as EM) (bioMerieux, Marcy I'Etoile, France) was the thirdmethod tested. Extractions were also performed with three commercialbead beating methods: the ZR Fecal DNA MiniPrepTM (ZR) (ZymoResearch, Irvine,CA, USA), the FastDNA@ SPIN Kit for Feces (FpF)(MP Biomedicals, Santa Ana, CA, USA), and the FastDNAQ SPIN Kit forSoil (FpS)(MP Biomedicals). Bead beating was performed withthe FastPrep@-24 Instrument (MP Biomedicals) at 6.0 msfor 40 s.Commercial methods were used as recommended by the manufacturerswith slight modifications for MG + and QS. For MG +, an additionalphenol-chloroform purification step was added before DNA precipita-tion with isopropanol to increase the purity of the recoveredDNA incomparison to the manufacturer's protocol (A260/A280 =1.6±0.5versus 1.4±0.5). For QS, we added, as recommended by several au-thors (Claassen et al., 2013; Smith et al, 2011), an initial step of 1-minvortexing at 30 Hz in ASL buffer, and used the manufacturer's optionalpre-treatment step of heating samples in a water bath at 95℃ for10 min. The non-commercial DNA extraction method is an SDS gentlelysis-based protocol (BO) developed by Porteous et al.(1997), later onmodified by Bonot et al. (2010). These modifications included the con-ditioning of the sludge samples as a liquor, and the use of reduced con-centrations of guanidine isothiocyanate, which limits the risk ofcarrying over this chaotropic agent in the DNA extracts,as explainedin detail by the authors. This method was chosen because of its versatil-ity in extracting good quality DNAs from environmental matrices as dif-ferent as soil/sediment and activated sludge (Bonot et al.,2010). 2.4. DNA quantification and quality testing DNA yield was assessed using the Qubit@ dsDNA HS Assay kit with aQubit@ 2.0 fluorometer (Life Technologies,Carlsbad,CA, USA). The pu-rity of DNA was expressed as the ratio of absorbance at 260 nm and280 nm (A260/A280) using a Nanodrop@ spectrophotometer (NanoDropTechnologies, Wilmington,DE,USA). DNA shearing was evaluated byelectrophoresis in 0.8%agarose/TBE (Tris-borate-EDTA buffer) gel con-taining 1x GelRedTM dye (Biotium, San Francisco, CA, USA). The DNAwas visualized using the Molecular Imager@ Gel DocTM XR System(Bio-Rad Laboratories, Inc., Hercules, CA,USA). 2.5. PCR inhibition testing The absence of residual inhibitors in fecal DNA extracts was evaluat-ed by analyzing samples that were spiked with a known amount ofpB10 plasmid using a pB10-specific qPCR assay. In these tests, any tracesof inhibitors were expected to reduce the expected quantity of pB10copies detected by qPCR, compared to a control containing pB10 with-out fecal DNA. The plasmid pB10 was extracted from overnight culturesof E. coli DH5a(pB10) in Lysogeny Broth using the QIAGEN@ PlasmidMidi Kit (Qiagen).The concentration and purity of plasmid DNA were Extraction method (abbreviation) Starting amount (mg) Cell lysis procedure Average cost"(E/sample) Completion time MasterPureTM Gram Positive (MG +) ND L,CLB 2.09 24h QIAamp@ DNA Stool (QS) 220 CLB.T 436 2h NucliSENSO easyMAG@ (EM) S200 CLB 9.58 1 h 15 min Bonot et al. (2010) (BO) ≤500 SDS,US <2 48h ZR Fecal DNA MiniPrepTM (ZR) ≤150 BB,CLB 4.54 1h FastDNA@ SPIN Kit for Feces (FpF) ≤500 BB.CLB 7.72 1h 15 min FastDNA@ SPIN Kit for Soil (FpS) ≤500 BB.CLB 7.72 1 h 30 min If specified by the manufacturer,L: lysozyme,T: temperature, CLB: cell lysis buffer, SDS: sodium dodecyl sulfate, US: sonication, BB: bead beating. ”Prices as of November 2013. determined spectrophotometrically based on absorbance readings at260 and 280 nm, using a Nanodrop@ spectrophotometer (Nanodroptechnologies). A260 was used for the concentration calculation whilethe ratioA260/A280 was used for estimation of purity. Fecal DNA samplesof 10 and 25 ng were spiked with pB10 plasmid DNA (10°copies ofpB10 per reaction). Spiked DNA samples were used as templates inqPCR targeting pB10. PCR assays were performed with the MyiQTM2real-time PCR system (Bio-Rad Laboratories) using pB10-specificprimers (Bonot and Merlin, 2010) and MESA FAST qPCR MasterMixfor SYBR@Assay as recommended by the manufacturer (Eurogentec,Seraing, Belgium). All assays were performed in triplicate. The followingthermocycling conditions were utilized: initial denaturation at 95 C for5 min, 40 cycles of 95℃ for 15 s, 60°℃for 1 min, and a final extension at72 C for 5 min. Melting curves were obtained immediately after theamplification under the following conditions: denaturation at 95Cfor 1 min,annealing at 60 ℃ for 1 min,and 70 cycles of 10 s with an in-crement of 0.5C/cycle starting at 60C. The specificity of the primersets for detecting pB10 was systematically checked by verifying theabsence of a qPCR amplification signal (detection limit =3 copies perreaction) using fecal DNA before spiking (control). 2.6. Total 16S rRNA gene quantification The total bacteria amount was assessed by amplifying 0.5 ng of eachfecal extract with pan-bacterial primers targeting the 16S rRNA gene:Eub338F (5'-ACTCCTACGGGAGGCAGCAG-3') and Eub518R (5'-ATTACCGCGGCTGCTGG-3') (Guo et al.,2008). The reaction mix and thethermocycling conditions were identical to those used for pB10 amplifi-cation. Each sample was tested in triplicate. Standard curves rangingfrom 10 to 10 copies of 16S rDNA per reaction were obtained usingtenfold serial dilutions of about 1500 bp-amplicons of 16S rDNA obtain-ed from B. thetaiotaomicron ATCC 29741. B. thetaiotaomicron DNA wasextracted with the QIAamp@ DNA Mini Kit (Qiagen) and the amplifica-tion of its 16S rDNA was performed using primers P8 (5'-AGAGTTTGATCCTGGCTCAG-3') and P1525 (5'-AAGGAGGTGATCCAGCCGCA-3')(Edwards et al.,1989). The reaction mixture (50 pl total volume) foreach reaction was x1 reaction buffer (Invitrogen, Thermo FisherScientific, Waltham, MA, USA), 2 U Taq polymerase (Invitrogen),40 pmol of each primer, 2 mM MgCl2, 0.2 mM dNTPs and 2 plof thefinal DNA eluate. The following thermocycling conditions were utilized:initial DNA denaturation step at 95 ℃ for 5 min, 30 cycles of 95 ℃ for1 min, 65 C for 1 min, 72 C for 1 min, and a final extension at 72℃for 5 min. 2.7.Extraction efficiency of genomic DNA from bacteria with different cellwalls For each extraction method studied, lysis efficiency was evaluatedby testing recovery of known bacterial DNA from samples previouslyspiked with a known amount of a Gram-negative bacterium with athin cell wall (P. baroniae DSMZ 16972"), a Gram-positive bacteriumwith a thick cell wall (E. faecium BM4339), and a bacterium with amycolic acid-containing cell wall (M. chelonae CIP104535). Bacteriawere harvested from agar plates and suspended in sterile, filtered PBSin order to obtain an optical density corresponding to 0.5 McFarland (~1.5x10°bacteria·ml). The aliquots (50 mg) of feces and cecalcontents were then spiked with 650 ul of each strain suspension. Sam-ples were further centrifuged for 10 min at 13,000xg. The supernatantwas discarded and the pellet was used for DNA extraction. The amountsof spiked bacteria were confirmed by standard quantitative culture. Theextracted DNA was analyzed by strain-specific qPCR targeting chromo-somal genes that are present as single copies and are specific for eachtested organism (Table2). For each of these genes, the absence of thecorresponding sequences in mice cecal contents and feces was verified(limit of detection of each gene = 5 copies per reaction).PCR assayswere performed in triplicate on 5 ng of fecal or cecal DNA with theMyiQTM2 real-time PCR system (Bio-Rad Laboratories) using specificprimers (Table 2) and MESA FAST qPCR MasterMix for SYBR@Assay(Eurogentec). The thermocycling conditions were identical to thoseused for pB10 amplification. The standard curves ranging from 10 to10’ copies per reaction were constructed based on DNA extractedfrom the respective bacterial strains using the QIAamp@ DNA Mini Kit(Qiagen). 2.8. Statistical analysis Statistical analyses were performed using the Minitab@ 16.2.3 Sta-tistical Software (Minitab, Inc., State College,PA,USA). Analyses weredone with log transformed data. Data were expressed by means ±standard error of the means. Normality for all data sets was testedusing the Anderson Darling normality test. Multiple comparisons wereconducted using Welch's ANOVA (one-way analysis of variance forheteroscedastic data). A p-value of less than 0.05 was considered statis-tically significant. 3. Results 3.1. Total DNA yield, purity and integrity Total DNAs were extracted from mice fecal and cecal samples usingeach of the seven extraction methods listed in Table 1. For commercialmethods, different amounts of starting material are recommended bythe manufacturers. In this study, DNAs were extracted from 50 mg ofstarting material (wet weight). This amount was chosen since prelimi-nary experiments (performed using 50, 100, 150, and 200 mg samples)showed that there was a negative relation between the amount of fecalmaterial and DNA yield for three methods (MG+, FpF and FpS) forwhich samples≥100 mg produced impure and sheared DNA (datanot shown). The mean DNA yields, determined by fluorometry, varied greatly ac-cording to the methods used for extraction (Table 3). For feces, thehighest DNA yields (gsample) were obtained with the BO method(mean, 134 pg/g) and the three bead beating kits (FpS, FpF and ZR)(mean range,58.02-64.20 pg/g). For cecal samples, the highest amountsof DNA were obtained with the FpS kit (mean, 74.55 pg/g)and, to a lesserextent,with the FpF kit (mean,36.64pg/g).DNA yields obtained for cecalsamples with the other methods were much lower (mean range, 5.09-7.83 ug/g). For fecal and cecal samples, the purity of extracted DNA,assessed by determining the A260/A280 ratio (optimal ratio comprisedbetween 1.8 and 2), was satisfactory with FpS, EM and BO (Table 3). Strain Target gene Primers (5'→3') Reference Prevotella baroniae niml ATGTICAGAGAAATGCGGCGTAAGCGGCTTCCTTGCCTGTCATGTGCTC Trinh et al.(1996) DSMZ 16972 Mycobacterium chelonae IIs GTTTCTGTAGTGGTTACTCGCTTCAGCTCCCCGAGGCWTATC Modified from Ngan et al.(2011) CIP 104535 Enterococcus faecium BM4339 vanD TAAGGCGCTTGCATATACCGTGCAGCCAAGTATCCGGTAA Perichon et al. (1997) Yields and purity of DNA extracted from mice feces and cecal contents with seven com-pared methods. Extraction method Nucleic acid yield A260/A280 16S rRNA gene yield (ng/g sample)" (log copies/g sample) Feces MG+ 25.90土10.70 1.62±0.05 10.64±0.15 35.47土1.39 2.10±0.01 11.15±0.02 19.66±9.54 1.99±0.02 10.75±0.24 134.00土15.80 1.93±0.03 12.02土0.10° 64.20土24.20 1.600.54 11.41±0.38 58.027.60 1.92±0.02 11.97±0.04 63.6915.13 2.230.03 11.81±0.04" Cecal contents MG+ 6.83±1.46 1.95±0.04 10.54土0.07 5.090.96 2.16±0.05 10.30±0.13 7.8311.95 1.930.01 10.83±0.11 6.34±2.37 1.79土0.10 10.75±0.23 7.04±5.11 1.23斗0.58 10.73±0.17 74.55±1.59 1.94±0.01 11.94±0.06° 36.64±8.63 2.98±0.14 11.30土0.02 MG+:MasterPureTM Gram Positive; QS: QIAamp@ DNA Stool; EM: NucliSENSeasyMAG@; BO: method from Bonot et al. (2010); ZR: ZR Fecal DNA MiniPrepM; FpF: FastDNA@ SPIN Kit for Feces; FpS: FastDNA@ SPIN Kit for Soil "DNAyield from a 50 mg sample as determined with the Qubit fluorometer (averageoffive replicates 土 SEM). " Significantly different from the other four or six methods, respectively (p<0,001). The lowest A260/A280 ratio was observed in the ZR extracts obtained fromfecal samples (mean ratio,1.60) and cecal samples (mean ratio, 1.23).The purity of the extracts obtained using the MG+ kit was unsatisfacto-ry (mean ratio, 1.62) for feces. For extracts obtained from fecal and cecalsamples using the QS kit, and extracts obtained from fecal samples usingthe FpF kit, the A260/A2s0 ratio was above 2. The quality of the recovered DNA was assessed in terms of DNAfragmentation by agarose gel electrophoresis (Fig.1). With the FpS kit,most of the DNA recovered from fecal or cecal samples was preservedwith a majority of DNA fragments being>5 kb. Similar results wereobserved for extracts obtained from cecal contents with the FpF andBO methods. However, with these methods, DNA from fecal extractswas overall more degraded, particularly with the BO method, whichalso yielded large quantities of sheared DNA with low molecular weight.For each type of samples, DNA obtained with the EM and ZR kits, waspredominantly smeared while some intact bands >5 kb were observedwith MG + extracts. 3.2. Presence of PCR inhibitors We tested the DNA extracted with the seven methods for the pres-ence of PCR inhibitors by mixing a known amount of pB10 plasmid (10°copies per reaction) with 10 or 25 ng of extracted DNA. Nodetectable inhibition could be demonstrated with 10 ng of DNA extractswhatever the extraction method was. When 25 ng DNA extracts weretested, a detectable inhibition was found only in the case of theMG + kit (inhibition ranging from 13% up to 90%, depending on thereplicate). Thus, no inhibitors were detected in the applied templaterange used in our qPCR assays. 3.3. Extraction efficiency of bacterial DNA The quantity of total bacterial DNA content is an important criterionfor evaluating DNA extraction methods as it assesses the representative-ness of bacterial populations. The number of 16S rRNA gene copies perstarting material (gof samples) was thus quantified by qPCR. Thisnumber varied up to 10- to 100-fold between the methods rangingapproximately from 10 to 12 log (Table 3). For feces,significantly higheramounts of 16S rDNA were obtained with BO, FpS and FpF (p<0.001).Amounts of 16S rDNA obtained from cecal contents were significantlyhigher only when the FpS kit was used (p<0.001). The capacity to lyse different types of bacterial wall was assessed byadding three different model strains in the sample prior to extractionand quantifying back the bacteria by qPCR from the DNA extractsusing specific targets (niml for P. baroniae DSMZ 16972, vanD forE. faecium BM4339 and ITS for M. chelonae CIP 104535). Among allmethods tested, the FpS kit yielded the highest recovery rate (approxi-mately the totality of the initial inoculum per 50 mg sample i.e.approx-imately 10°CFU of P. baroniae, 10°CFU of E. faecium and 10CFU ofM. chelonae) for all three strains whether tested in feces or cecalcontents (Fig.2). The two other bead beating methods (FpF and ZR)were significantly less efficient (p <0.001) for extracting DNA fromE. faecium BM4339 (revovery of vanD: 60% with FpF and 2% with ZR)and M. chelonae CIP 104535 (recovery of ITS: 25% with FpF and 10%with ZR). Less than 1% of the expected amounts of mycobacterial andenterococcal target genes were obtained with the other four extractionsmethods whether tested on feces or cecal contents. 4. Discussion The purpose of this study was to select an optimal DNA extractionmethod that may be used for molecular assays,particularly qPCRbased experiments, aimed at detecting and characterizing bacterialgenes in intestinal samples from mice. It is noteworthy that preliminaryexperiments aiming at adjusting the optimal quantities of startingmaterial showed that excess amount of feces had negative effects onthe output of the extraction, as more DNA fragmentation and impuritieswere observed. This is reminiscent to what was reported by Ariefdjohanet al.(2010) with human feces while using commercial kits includingFast DNA@ SPIN Kit for Soil. This effect may be due to an overloading Fig. 2. Log10 copies of detected niml, vanD and ITS sequences (50 mgof feces or cecal contents). Mean values are indicated by the line within the box plot. The box extends from the 25thto 75th percentiles and whiskers indicate the minimum and maximum values. Significantly different from BO and EM (p<0.001). "Significantly different from the other six methods(p<0.001). Significantly different from MG+,QS, EM and BO (p<0.001). "Significantly different from MG+, QS,EM, BO and ZR (p<0.001). MG+: MasterPureM Gram Positive; QS:QIAamp@DNA Stool; EM: NucliSENS@ casyMAG@; BO: method from Bonot et aL(2010);ZR: ZR Fecal DNA MiniPrepTM;FpF: FastDNA@ SPIN Kit for Feces; FpS: FastDNA@ SPIN Kit for Soil. of the purification matrix. This clearly points out the importance ofadjusting the amount of starting material beforehand for DNA extrac-tion when sensitive molecular techniques such as qPCR are expectedto be used. The relatively high dietary fiber content of mice feces is known as afactor that may interfere with PCR assays (Monteiro et al., 1997).However, in our study, all methods tested were found to ensure efficientremoval of PCR inhibitors regardless of the nature of the samples tested.We found that extraction methods based on the use of mechanicaltreatment of samples, i.e. bead beating (FpS, FpF, ZR) or sonication(BO), produced higher amounts of DNA from feces than purely chemicalmethods (MG+, QS, EM). The positive impact of mechanical disruptionwas also observed for feces in terms of detection of 16S rDNA since sig-nificantly higher concentrations of 16S rRNA gene copies were detectedin extracts obtained using the FpS, the FpFand the BO methods. It is pos-sible that this impact is particularly important for mice feces because oftheir dryness.Nevertheless, it is unclear whether this effect was greater in mice than in other animals or in humans. Indeed, if the positive im-pact of mechanical treatments was also noted for human feces or sludgespecimens (Ariefdjohan et al., 2010; Claassen et al., 2013; Guo andZhang, 2013; Salonen et al., 2010), it is difficult to compare the resultsobtained in these studies with ours because of methodological differ-ences such as fecal sample size, methods used to quantify total and bac-terial DNA,and extraction protocols. Feces differ in their bio-structure from intestinal contents (Pang et al.,2012; Gu et al.,2013). Thus, an optimal method should allow an equallyefficient extraction from both fecal and intra-intestinal materials to en-sure unbiased comparisons. Higher yields of total DNA and 16S rDNAwere also obtained when mechanical treatments were applied to cecalsamples. However, in this case, significantly higher extraction efficien-cies were achieved with the FpS and FpF kits only. These results are, atleast partially, in accordance with those of Guo and Zhang (2013) whofound that the FpS kit was more efficient than the other methods tested,including QS, for DNA extraction of sludge samples. It has been reported that bead beating may cause DNA degradationrendering it unsuitable for various downstream applications such asPCR, especially in assays targeting relatively large sequences (Olsonand Morrow, 2012). In our study, the integrity of DNA varied dependingon the sample types and extraction methods used. Bead beating causedsome shearing but, except for the ZR method, the majority of DNAfragments recovered with these methods were larger than 1.6 kb, thelength of entire rrs genes, making them suitable for PCR-analyses ofmicrobiomes. These results are consistent with other studies showingthat good quality DNA may be obtained from sludge or human intestinalsamples using bead beating methods (Ariefdjohan et al., 2010; Guo andZhang, 2013; Salonen et al.,2010). Lower sensitivity of PCR detection in various specimens, includingfeces, of genes present in Firmicutes and Actinobacteria may also berelated to the use of DNA extraction methods that are insufficient to pro-vide efficient cell wall lysis (Maukonen et al., 2012). Bead beatingmethods have been reported to be more efficient than chemical methodsin detecting genes of various Gram-positive bacteria present in humanfecal samples and sludge samples (Guo and Zhang, 2013; Salonen et al.,2010). Consistent with these results, we found that among all extractionmethods tested, the FpS kit was the most efficient for extracting DNAfrom E. faecium regardless of the nature of the samples tested. This kitwas also the most efficient for Mnnnnn chelonae DNA extraction inboth fecal and cecal matrices. These findings are in accordance withthose reported by Pontiroli et al. (2011) who evaluated the efficienciesof different DNA extraction methods for the detection by PCR of Myco-bacterium bovis in bovine fecal samples. In this study, FpS performed bet-ter than purely chemical methods and was more efficient than the QSmethod used alone or in combination with a bead beating step. It is worth noting that DNA extraction can be time consuming, whichconstitutes often a bottleneck of various molecular analyses. Althoughmore expensive, all commercial kits allowed us to considerably reducethe processing time as compared with the cheapest non-commercialBO method which necessitates 48 h of manipulations. 5. Conclusion To the best of our knowledge, this is the first study that comparesDNA extraction methods applied to feces and intestinal contents ofmice. Of the seven methods tested, the FpS method was the mostefficient for extracting bacterial DNA of good quality from feces andintra-intestinal contents of mice. With the other two bead beatingbased kits (FpF, ZR) and the BO method, DNA yield, DNA quality andcell wall lysis efficiency were more variable depending on the natureof specimens tested. Overall the worst performances were observedwith the three purely enzymatic kits tested, including QS, which is themost currently used extraction kit for fecal samples. Thus, we recom-mend the FpS kit to extract qPCR-grade DNA from mice fecal samples. Potential conflicts ofinterests Alain Lozniewski is a member of the Scientific Board of bioMerieux.France. Acknowledgments The authors sincerely thank Elodie Naguel and Agathe Inigo for theirtechnical assistance. References Alexander, T.W.,Yanke, j.L,Reuter, T.,Topp, E.,Read, R.R.,Selinger, B.L,McAllister, T.A.,2011. Longitudinal characterization of antimicrobial resistance genes in feces shedfrom cattle fed different subtherapeutic antibiotics. BMC Microbiol. 11,19. Ariefdjohan, M.W.,Savaiano, D.A.,Nakatsu, C.H, 2010. Comparison of DNA extraction kitsfor PCR-DGGE analysis of human intestinal microbial communities from fecalspecimens. Nutr.j.9,23. Bellanger, X.,Guilloteau, H.,Bonot, S.,Merlin, C., 2014. Demonstrating plasmid-basedhorizontal gene transfer in complex environmental matrices: a practical approachfor a critical review. Sc. Total Environ. 493C. 872-882. Bonot,S.,Merlin, C., 2010.Monitoring the dissemination of the broad-host-range plasmidpB10 in sediment microcosms by quantitative PCR. Appl. Environ. Microbiol. 76,378-382. Bonot, S,Courtois, S.,Block,J.-C.,Merlin, C, 2010. Improving the recovery of qPCR-gradeDNA from sludge and sediment. Appl. Microbiol. Biotechnol. 87, 2303-2311. Claassen,S.,du Toit, E.,Kaba,M.,Moodley,C,Zar,H.J,Nicol, M.P., 2013.A comparisonof theefficiency of five different commercial DNA extraction kits for extraction of DNA fromfaecal samples. j. Microbiol. Methods 94, 103-110. Dundas, N.,Leos, N.K.,Mitui, M.,Revell,P.,Rogers, B.B., 2008. Comparison of automatednucleic acid extraction methods with manual extraction. j.Mol. Diagn.10,311-316. Edwards, U.,Rogall, T., Blocker, H.,Emde, M., Bottger, E.C., 1989. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene codingfor 16S ribosomal RNA. Nucleic Acids Res. 17, 7843-7853. Gu, S.,Chen,D.,Zhang, J.N.,Lv, X,Wang, K.,Duan, LP.,Nie, Y.,Wu, X.L, 2013. Bacterialcommunity mapping of the mouse gastrointestinal tract. PLoS One 8, e74957.Guo,F,Zhang,T.,2013. Biases during DNA extraction of activated sludge samples revealed by high throughput sequencing. Appl. Microbiol. Biotechnol.97,4607-4616. Guo,X,Xia,X.,Tang,R,Zhou,j.,Zhao,H.,Wang, K.,2008. Development of a real-time PCRmethod for Firmicutes and Bacteroidetes in faeces and its application to quantify intes-tinal population of obese and lean pigs. Lett. Appl. Microbiol. 47, 367-373. Inglis, G.D,Thomas, M.C.,Thomas, D.K,Kalmokoff, M.L,Brooks, S.P.,Selinger, LB., 2012.Molecular methods to measure intestinal bacteria: a review.J. AOAC Int. 95,5-23. Li, M.,Gong,J.,Cottrill, M.,Yu, H., de Lange, C., Burton, J., Topp, E., 2003. Evaluation ofQIAamp@ DNA Stool Mini Kit for ecological studies of gut microbiota. j Microbiol.Methods 54. 13-20. Maukonen,J.,Simoes, C.,Saarela,M., 2012. The currently used commercial DNA-extractionmethods give different results of clostridial and actinobacterial populations derivedfrom human fecal samples. FEMS Microbiol. Ecol. 79,697-708. Monteiro, L,Bonnemaison,D.,Vekris, A.,Petry, K.G.,Bonnet,」.,Vidal,R.,Cabrita,J.,Megraud,F, 1997. Complex polysaccharides as PCR inhibitors in feces: Helicobacter pylorimodel. j.Clin. Microbiol. 35,995-998. Ngan,G.J..Ng, L.M., Jureen,R.,Lin, R.T.,Teo,J.W., 2011. Development of multiplex PCRassays based on the 16S-23S rRNA internal transcribed spacer for the detection ofclinically relevant nontuberculous mycobacteria. Lett. Appl. Microbiol. 52, 546-554. Nylund, L,Heilig, H.G.,Salminen,S.,de Vos, W.M.,Satokari, R., 2010. Semi-automatedextraction of microbial DNA from feces for qPCR and phylogenetic microarrayanalysis.j. Microbiol. Methods 83.231-235. Olson, N.D.,Morrow, J.B., 2012. DNA extract characterization process for microbialdetection methods development and validation.BMC Res. Notes 5,668. Pang. W.,Vogensen, F.K.,Nielsen,D.S.,Hansen, A.K., 2012. Faecal and caecal microbiota profiles of mice do not cluster in the same way. Lab.Anim. 46,231-236.Perichon, B.,Reynolds, P.,Courvalin,P, 1997.VanD-type glycopeptide-resistant Enterococcusfaecium BM4339.Antimicrob. Agents Chemother. 41, 2016-2018. Pontiroli, A.,Travis, E.R.,Sweeney, F.P.,Porter,D.,Gaze, W.H.,Mason, S.,Hibberd, V.,Holden,J,Courtenay,O.,Wellington, E.M.,2011.Pathogen quantitation in complex matrices: amulti-operator comparison of DNA extraction methods with a novel assessment ofPCR inhibition. PLoS One 6, e17916. Porteous, LA.,Seidler,R.J,Watrud, LS., 1997. An improved method for purifying DNAfrom soil for polymerase chain reaction amplification and molecular ecology applica-tions. Mol. Ecol. 6.787-791. Presley,LL,Wei, B.,Braun,J,Borneman,j., 2010. Bacteria associated with immunoregula-tory cells in mice. AppL Environ. Microbiol. 76,936-941. Rapp,D.,2010. DNA extraction from bovine faeces: current status and future trends.」.AppL Microbiol. 108,1485-1493. Salonen, A.,Nikkila,J.Jalanka-Tuovinen,J.,Immonen,O.,Rajilic-Stojanovic, M.,Kekkonen,R.A., Palva, A., de Vos, W.M., 2010. Comparative analysis of fecal DNA extractionFemethods with phylogenetic microarray:effective recovery of bacterial and archaealDNA using mechanical cell lysis. j. Microbiol. Methods 81, 127-134. Schluter, A, Heuer, H.,Szczepanowski,R.,Forney, Lj.,Thomas, C.M.,Puhler, A.,Top, E.M.,2003. The 64 508 bp IncP-1beta antibiotic multiresistance plasmid pB10 isolatedfrom a waste-water treatment plant provides evidence for recombination betweenmembers of different branches of the IncP-1beta group.Microbiology 149,3139-3153. Scupham, A.JJones, J.A.,Wesley,LV.,2007. Comparison of DNA extraction methods for)am_analysis of turkey cecal microbiota. j.Appl. Microbiol. 102, 401-409. Smith, B.,Li, N.,Andersen, A.S.,Slotved, H.C.,Krogfelt, K.A., 2011.Optimising bacterial DNAextraction from faecal samples: comparison of three methods.Open Microbiol.j.5,14-17. Tang.jN.,Zeng,ZG.,Wang, H.N.,Yang,T.,Zhang,P.J..Li,Y.L,Zhang, A.Y.,Fan, W.Q,Zhang,Y.,Yang,X.,Zhao, S.J.,Tian,G.B.,Zou, L.K.,2008. An effective method for isolation of DNAfrom pig faeces and comparison of five different methods. j.Microbiol. Methods 75,432-436. Thompson, C.L,Hofer,M.J, Campbell, LL,Holmes, A.J.. 2010. Community dynamics in themouse gut microbiota: a possible role for IRF9-regulated genes in communityhomeostasis. PLoS One 5,e10335. Trinh, S., Haggoud, A., Reysset, G., 1996. Conjugal transfer of the 5-nitroimidazoleresistance plasmid plP417 from Bacteroides vulgatus BV-17: characterization andnucleotide sequence analysis of the mobilization region.j. Bacteriol.178,6671-6676. Turnbaugh, P.J.,Ley, R.E,Mahowald, M.A.,Magrini, V.,Mardis, E.R.,Gordon, j.1,2006. Anobesity-associated gut microbiome with increased capacity for energy harvest.Nature 444,1027-1031. Urdahl, A.M., Solheim,H.T., Vold, L,Hasseltvedt, V., Wasteson, Y., 2013. Shiga toxin-encoding genes (stx genes) in human fecal samples. APMIS 121, 202-210. http://dx.doi.org/j.mimet. Fig. .Electrophoretic profiles of DNA extracted from feces or cecal contents using the seven methods tested.MG+: MasterPureTM Gram Positive; QS:QIAamp@ DNA Stool; EM:NucliSENS@ easyMAGB; BO: method from Bonot et al.(; ZR: ZR Fecal DNA MiniPrepM; FpF: FastDNA@ SPIN Kit for Feces; FpS: FastDNA SPIN Kit for Soil. JaninaFerrand等人发表在Journal of Microbiological Methods杂志上的一篇文章中,对比了7种提取粪便DNA的方法。实验材料和方法1. 样品采集样本从五只8周的C57BL/6品系的小鼠中获得,有两种样本类型,分别为粪便样本和盲肠内容物。将粪便样本取出后立即放于-20℃条件下冻存。盲肠内容物样本经解剖得到后尽快放于-80℃冻存。2. DNA提取。所用不同方法或试剂盒种类如下:3.DNA纯度和质量检测。OD A260/A280比值测定以及0.8%TBE凝胶电泳实验。实验结果 图一 利用七种方法得到的DNA比较上图一可以看出,在针对粪便样本进行DNA提取中,利用FpS试剂盒得到的DNA得率为58.2μg/g,利用FpF试剂盒得到的DNA得率为63.69μg/g,与其他试剂盒相比DNA得率明显较高。另外,在针对盲肠内容物样本进行DNA提取过程中,利用FpS试剂盒得到的DNA得率为74.55μg/g,利用FpF试剂盒得到的DNA得率为36.64μg/g,与其他试剂盒以及提取方法(BO)相比,DNA得率遥遥领先,分别位于第一和第二位。 图二 利用七种方法得到的DNA电泳图结果比较从以上图二的电泳图可以看出,在粪便样本中,Fps泳道DNA无拖尾现象,表明DNA纯度较高,没有降解,且DNA片段大于5kb,证明提取得到的DNA完整性很高;在盲肠内容物样本中,Fps和FpF泳道无拖尾现象,无降解,且DNA片段大于5kb,证明得到的DNA纯度高,完整性高,可直接用于下游实验。实验结论经过以上7种实验方法的对比可以看出,不同的提取方法中样本的处方式不同,FpS,FpF方法使用了研磨珠与Fastprep-24 (MP Biomedicals)一起使用的方法;BO使用超声波来破碎样本;这些都属于使用仪器设备对样本进行物理破碎。显然通过物理破碎的方法比化学裂解的方法DNA得率高(如MG、QS、EM)。且只有FpS,FpF这两种方法得到的DNA质量和纯度较好
确定




还剩4页未读,是否继续阅读?
MP Biomedicals (Shanghai) Co.,Ltd. 安倍医疗器械贸易(上海)有限公司为您提供《粪便中DNA提取检测方案(研磨机)》,该方案主要用于医疗/卫生中DNA提取检测,参考标准--,《粪便中DNA提取检测方案(研磨机)》用到的仪器有MP Fastprep-24 5G 快速样品制备仪
推荐专场
相关方案
更多
















