方案详情
文
Results demonstrate that monolayer surfaces permit accurate SPR analysis of macromolecular interactions and that the SPRi-PLEX configuration is well suited to a simple recovery protocol that provides rapid sensitive mass spectrometry analysis. Clearly at these levels of surface saturation more than 100 spots could be rapidly visualized and the material recovered for analysis by SELDI even with the manual approach used here.
方案详情

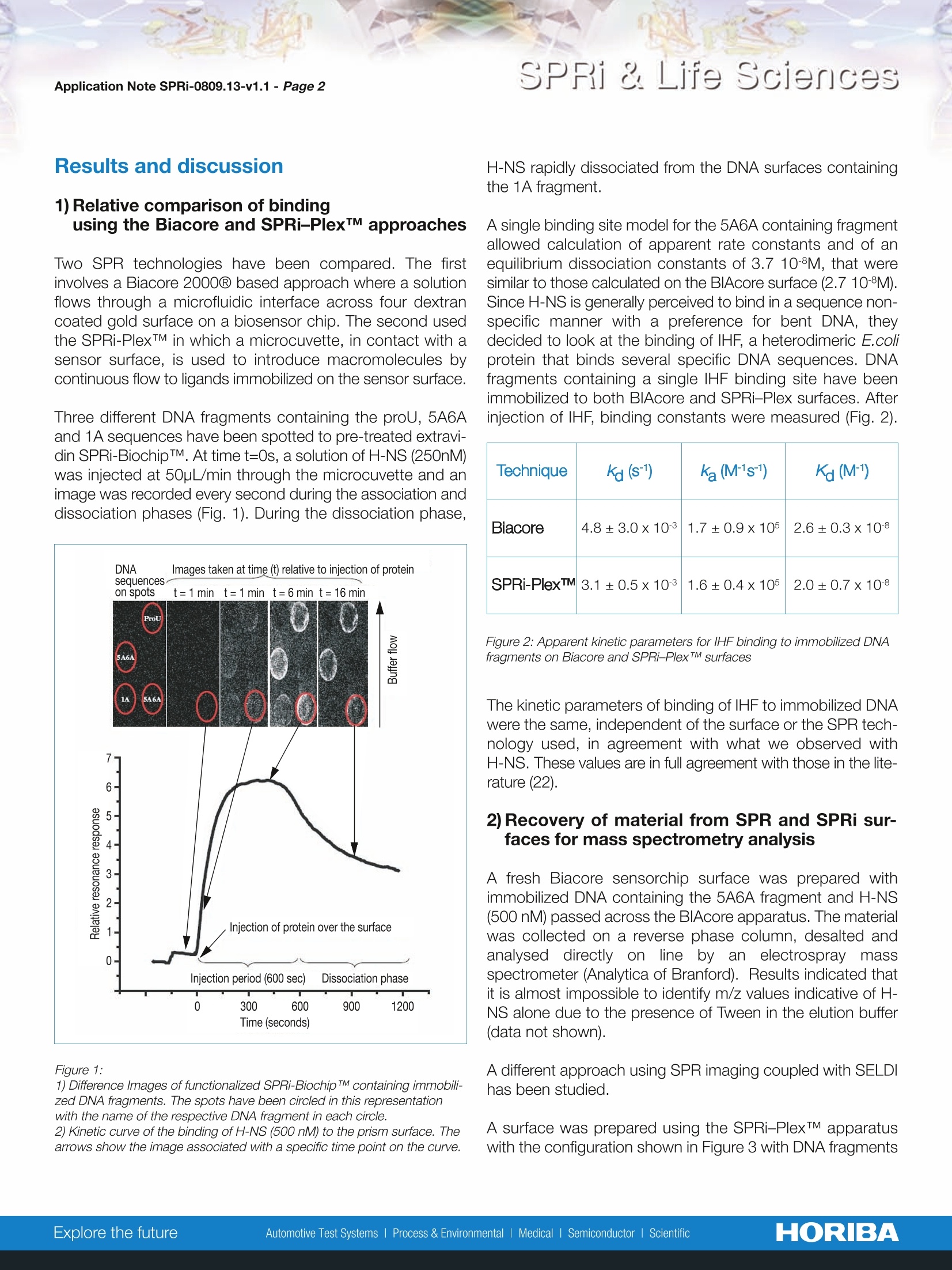
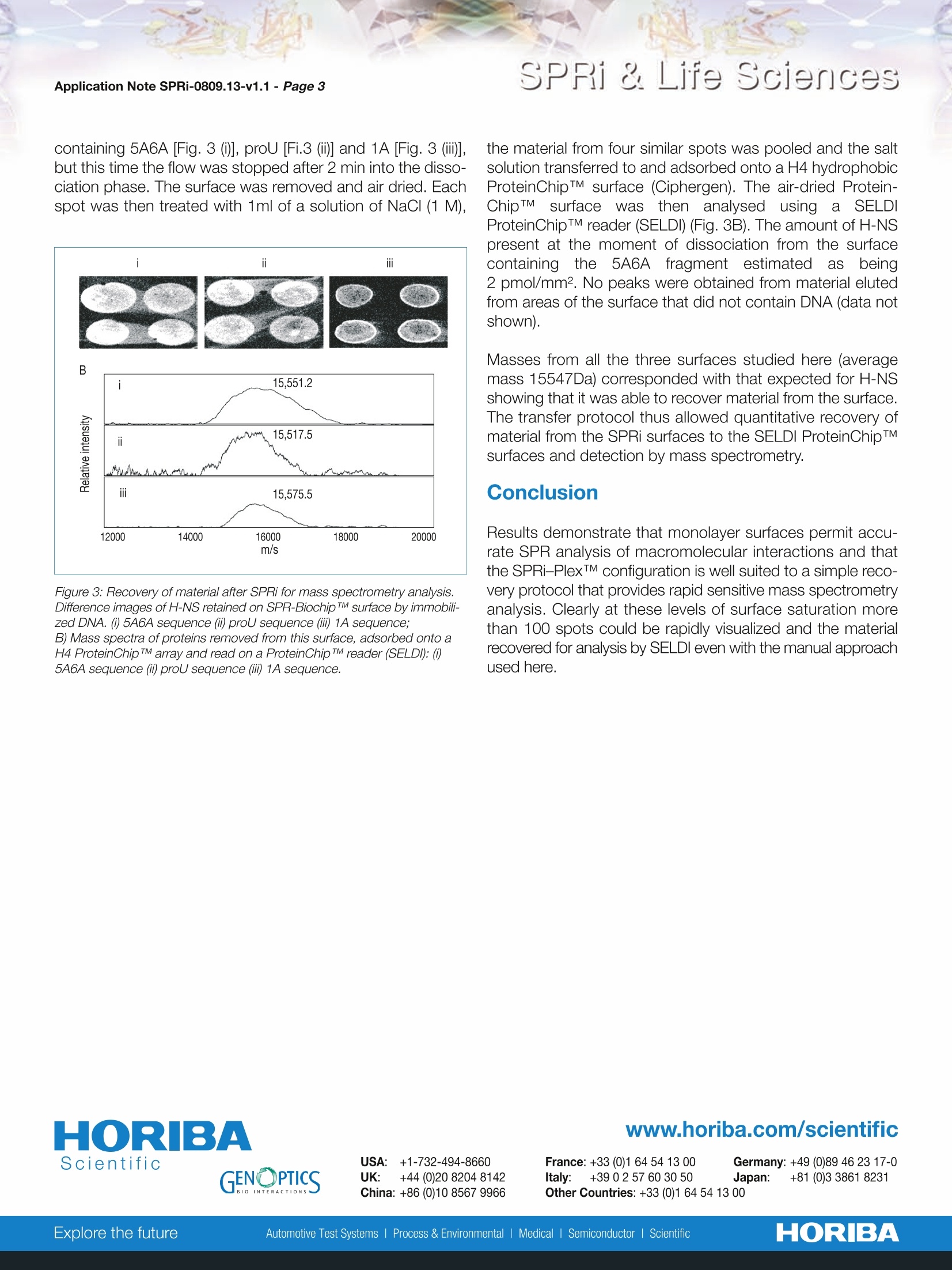
Scientific SPRi & Life SciencesApplication Note SPRi-0809.13-v1.1- Page 2 HORIBA SPRi & Life Sciences Rapid coupling of Surface Plasmon Resonance (SPR and SPRi) and ProteinChipTMbased mass spectrometry for the identification of proteinsin nucleoprotein interactions Emeline Bouffartigues, Herve Leh, Marielle Anger-Leroy, Sylvie Rimsky and Malcolm Buckle Nucleic Acids Research, 2007, Vol. 35, No. 6 The coupling approaches of SPR to LC-MS and ProteinChipTM-based mass spectrometry(SELDI) as a means of identifying proteins captured on DNA surfaces has been compared. Thepotential to allow multiple, quantitative analysis of macromolecular interactions followed byrapid mass spectrometry identification of retained material has been studied on the bacterialnucleoid protein H-NS. A number of H-NS responsive promoters have been shown to containregions of intrinsic DNA curvature located either upstream or downstream of the transcrip-tion start point. A working model for H-NS binding has been proposed invoking differentialbinding to relatively high and low-affinity sites. Consequently, differential H-NS binding toimmobilized DNA containing high and low-affinity sites has been studied by SPRi and, thenrecovered and identified by mass spectrometry. 1) Biotinylated DNA probes produced by PCR For H-NS: 一5A6A (223-bp fragment with 75-bp curved sequence) 1A (220-bp fragment non-curved sequence) -proU (372-bp fragment containing 200-bp region knownto bind H-NS). For lHF: -:205-bp fragment of the paca236. 2) Functionalization of SPRi-BiochipTMwith extravidin The gold surface of SPRi-BiochipTM was treated with 11-mercaptoundecanoic acid (MUA) coupled to a layer ofpoly(ethylenimine) to which extravidin was covalentlyattached. 3) Immobilization of biotinylated DNA probes Biotinylated DNA probes were spotted through the biotin-extravidin interaction. The singly 5'-end labeled DNAfragments containing the proU, 5A6A and 1A sequenceswere immobilized on extravidin functionalized SPRi-BiochipTMby direct spotting of 10pg/ml solutions of each DNA fragment. 4) SPRi experiments Spotted SPRi-BiochipsTM were inserted in the SPRi-PlexTMapparatus and washed by a short (10pL) injection of 1M NaClfollowed by continuous flow (50pL/min) with running buffer.Various concentrations of protein were injected across thesurface. The surface was regenerated with an injection of10pL of NaCI (1M) which removed any bound H-NS. H-NS(250 nM) was then reinjected at 50pL/min and after 2 mininto the dissociation phase the flow was stopped, the surfaceremoved from the apparatus and air dried. 5) iProtein recovery from the SPRi-PlexTM and SELDImeasurements Each spot was treated by the addition of 1 pL of NaCl (1M).The spots were allowed to sit for 30s then the solutions reco-vered and spotted onto a ProteinChipTM H50 hydrophobicsurface and left to incubate for 1h in a humid chamber toavoid evaporation. Excess liquid was removed from each ofthe spots which were then washed by the 2-fold addition of1pLof distilled water. The spots were then dried, co-crystal-lized with 1pL of a 20% solution of matrix and read in aSELDI ProteinChipTM reader. These same samples have been studied using the Biacore2000Q instrument and protein recovery has been analysedon LC-MS instrument (data not shown). Results and discussion 1) Relative comparison of bindingusing the Biacore and SPRi-PlexTM approaches Two SPR technologies have been compared. The firstinvolves a Biacore 2000 based approach where a solutionflows through a microfluidic interface across four dextrancoated gold surface on a biosensor chip. The second usedthe SPRi-PlexTM in which a microcuvette, in contact with asensor surface, is used to introduce macromolecules bycontinuous flow to ligands immobilized on the sensor surface. Three different DNA fragments containing the proU, 5A6Aand 1A sequences have been spotted to pre-treated extravi-din SPRi-BiochipTM. At time t=0s, a solution of H-NS (250nM)was injected at 50pL/min through the microcuvette and animage was recorded every second during the association anddissociation phases (Fig. 1). During the dissociation phase, Figure 1: 1) Difference Images of functionalized SPRi-Biochip TM containing immobili-zed DNA fragments. The spots have been circled in this representationwith the name of the respective DNA fragment in each circle. 2) Kinetic curve of the binding of H-NS (500 nM) to the prism surface. Thearrows show the image associated with a specific time point on the curve. H-NS rapidly dissociated from the DNA surfaces containingthe 1A fragment. A single binding site model for the 5A6A containing fragmentallowed calculation of apparent rate constants and of anequilibrium dissociation constants of 3.7 10-8M, that weresimilar to those calculated on the BIAcore surface (2.7 10-8M).Since H-NS is generally perceived to bind in a sequence non-specific manner with a preference for bent DNA, theydecided to look at the binding of IHF, a heterodimeric E.coliprotein that binds several specific DNA sequences. DNAfragments containing a single IHF binding site have beenimmobilized to both BIAcore and SPRi-Plex surfaces. Afterinjection of IHF, binding constants were measured (Fig. 2). Technique ka (s-1) ka (M1s-1) Ka (M-1) Biacore 4.8±3.0x103 1.7±0.9x105 2.6±0.3x108 SPRi-PlexTM 3.1±0.5x10-3 1.6±0.4x105 2.0±0.7x108 Figure 2: Apparent kinetic parameters for IHF binding to immobilized DNAfragments on Biacore and SPRi-Plex TM surfaces The kinetic parameters of binding of IHF to immobilized DNAwere the same, independent of the surface or the SPR tech-nology used, in agreement with what we observed withH-NS. These values are in full agreement with those in the lite-rature (22). 2)Recovery of material from SPR and SPRi sur-faces for mass spectrometry analysis A fresh Biacore sensorchip surface was prepared withimmobilized DNA containing the 5A6A fragment and H-NS(500 nM) passed across the BIAcore apparatus. The materialwas collected on a reverse phase column, desalted andanalysed directly on by an electrospray massspectrometer (Analytica of Branford). Results indicated thatit is almost impossible to identify m/z values indicative of H-NS alone due to the presence of Tween in the elution buffer(data not shown). A different approach using SPR imaging coupled with SELDIhas been studied. A surface was prepared using the SPRi-PlexTM apparatuswith the configuration shown in Figure 3 with DNA fragments containing 5A6A [Fig. 3 ()], proU [Fi.3 (ii)] and 1A [Fig. 3 (iii)],but this time the flow was stopped after 2 min into the disso-ciation phase. The surface was removed and air dried. Eachspot was then treated with 1ml of a solution of NaCl (1 M), Figure 3: Recovery of material after SPRi for mass spectrometry analysis.Difference images of H-NS retained on SPR-Biochip TM surface by immobili-zed DNA. (i) 5A6A sequence (ii) proU sequence (iii) 1A sequence; B) Mass spectra of proteins removed from this surface, adsorbed onto aH4 ProteinChip TM array and read on a ProteinChip TM reader (SELDI:()5A6A sequence (i) proU sequence (iii) 1A sequence. the material from four similar spots was pooled and the saltsolution transferred to and adsorbed onto a H4 hydrophobicProteinChipTM surface (Ciphergen). The air-dried Protein-ChipTMsurfacewasthen analysed using a SELDIProteinChipTM reader (SELDI) (Fig. 3B). The amount of H-NSpresent at the moment of dissociation from the surfacecontaining the 5A6Afragment estimatedasbeing2 pmol/mm2. No peaks were obtained from material elutedfrom areas of the surface that did not contain DNA (data notshown). Masses from all the three surfaces studied here (averagemass 15547Da) corresponded with that expected for H-NSshowing that it was able to recover material from the surface.The transfer protocol thus allowed quantitative recovery ofmaterial from the SPRi surfaces to the SELDI ProteinChipTMsurfaces and detection by mass spectrometry. Results demonstrate that monolayer surfaces permit accu-rate SPR analysis of macromolecular interactions and thatthe SPRi-PlexTM configuration is well suited to a simple reco-very protocol that provides rapid sensitive mass spectrometryanalysis. Clearly at these levels of surface saturation morethan 100 spots could be rapidly visualized and the materialrecovered for analysis by SELDl even with the manual approachused here. France: +33 (0)1 64 54 13 00 Germany: +49 (0)89 46 23 17-0Italy: +39 0 2 57 60 30 50 Japan: +81 (0)3 3861 8231Other Countries: +33 (0)1 64 54 1300 HORIBAExplore the futureAutomotive Test Systems I Process & Environmental Medical SemiconductorlScientific Results demonstrate that monolayer surfaces permit accurate SPR analysis of macromolecular interactions and that the SPRi-PLEX configuration is well suited to a simple recovery protocol that provides rapid sensitive mass spectrometry analysis. Clearly at these levels of surface saturation more than 100 spots could be rapidly visualized and the material recovered for analysis by SELDI even with the manual approach used here.
确定

还剩1页未读,是否继续阅读?
HORIBA(中国)为您提供《蛋白质、生物大分子中相互作用,质谱分析检测方案(大分子作用仪)》,该方案主要用于其他中相互作用,质谱分析检测,参考标准--,《蛋白质、生物大分子中相互作用,质谱分析检测方案(大分子作用仪)》用到的仪器有HORIBA OpenPlex表面等离子体共振成像仪
推荐专场
相关方案
更多
该厂商其他方案
更多