方案详情
文
本研究旨在探讨缺氧诱导因子-1α (HIF-1α)和lncRNA 核富集丰富转录本1 (NEAT1)之间的关系,以及它们在缺氧条件下肝细胞癌(HCC)中的作用。NEAT1 和HIF-1α在肝细胞癌组织中呈高表达并呈正相关,其中NEAT1 高表达与肿瘤淋巴结转移(TNM)晚期及远处转移呈正相关; NEAT1 或HIF-1α上调的HCC 患者预后较差。NEAT1由HIF-1α诱导,siHIF-1α抑制NEAT1 表达;NEAT1 的过表达进一步促进了缺氧条件下肝癌的发展,同时促进细胞活力、迁移和侵袭,抑制细胞凋亡,下调HIF-1α可逆转这种作用。NEAT1 过表达促进肿瘤生长,下调HIF-1α可逆转NEAT1 促进肿瘤生长的发生, 揭示下调HIF-1α可抑制NEAT1 的表达,抑制HCC 的进展,改善预后。
方案详情

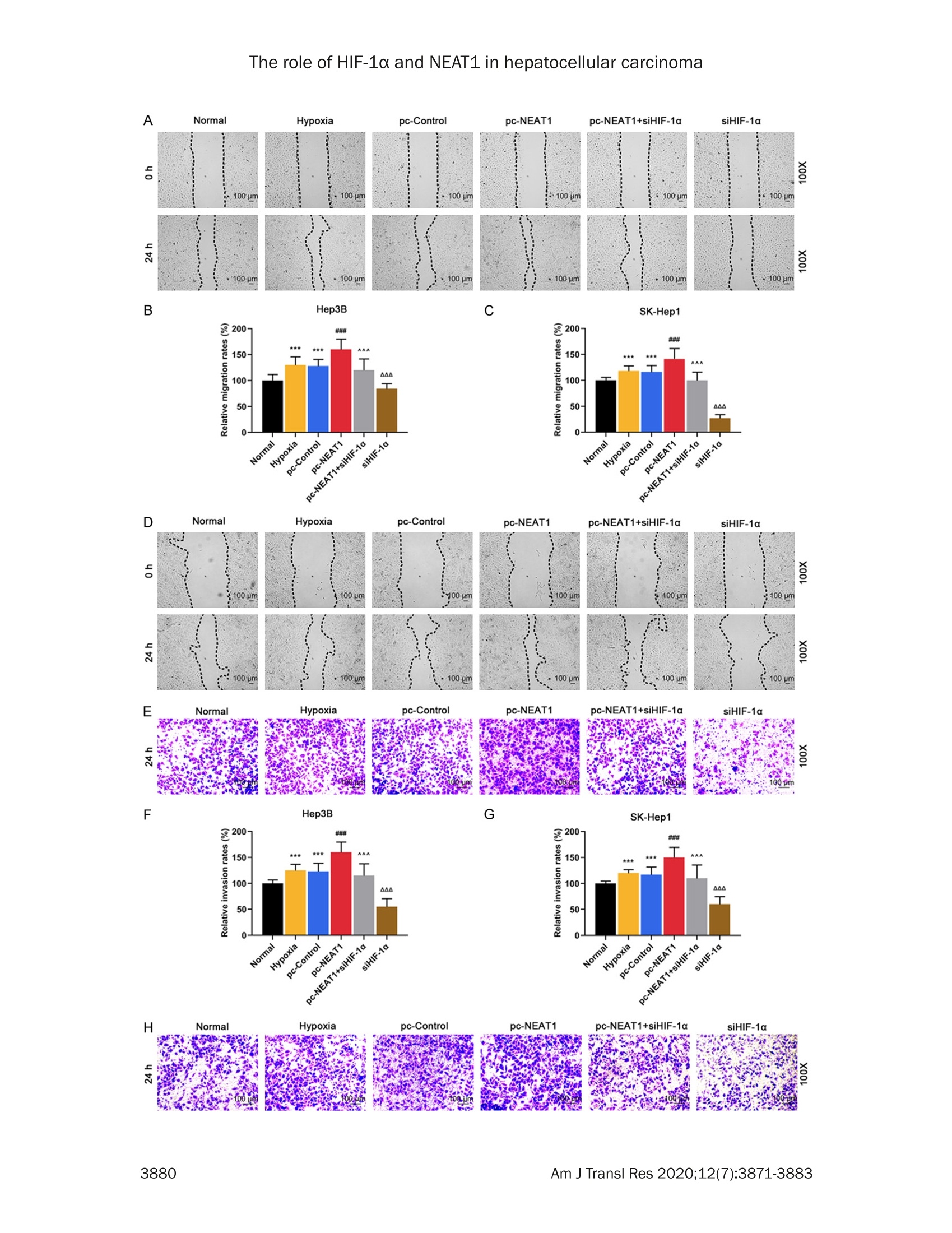
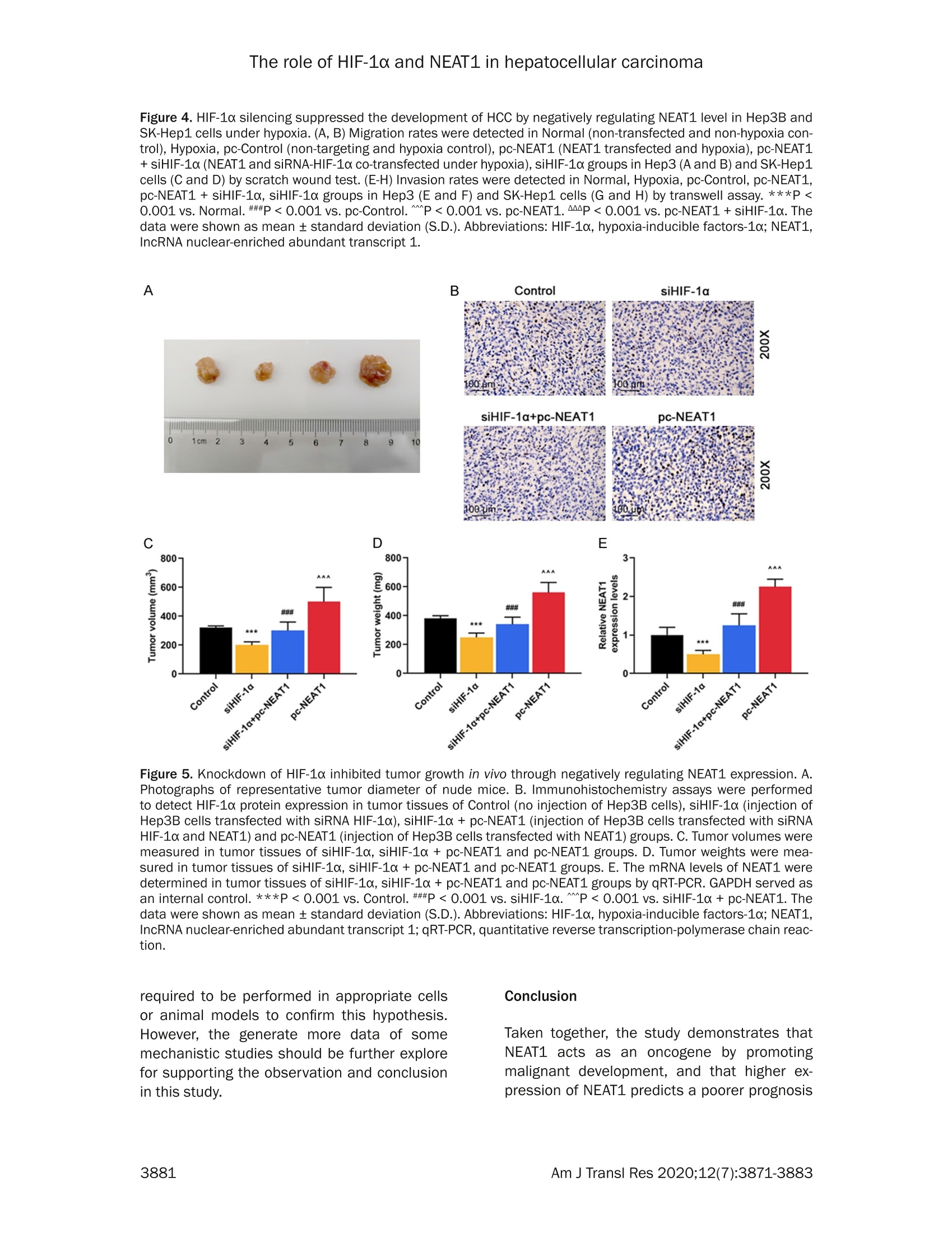
Am J Transl Res 2020;12(7):3871-3883www.ajtr.org/ISSN:1943-8141/AJTR0107148 The role of HIF-1a and NEAT1 in hepatocellular carcinoma Original Article Silencing of HIF-1a inhibited the expression of IncRNANEAT1 to suppress development of hepatocellularcarcinoma under hypoxia Xiuming Zhang1*, Zheng Kang1*, Xiaodong Xie1, Wei Qiao1, Lei Zhang1, Zhen Gong, Yan Chen1, WenrongShen1 1Department of Radiology, Jiangsu Cancer Hospital, Jiangsu Institute of Cancer Research, The Affiliated CancerHospital of Nanjing Medical University, Nanjing, China; 2Department of Gynecology, Women’s Hospital of NanjingMedical University, Nanjing Maternity and Child Health Care Hospital, Nanjing, China. *Equal contributors. Received December 30,2019; Accepted June 2, 2020; Epub July 15, 2020; Published July 30, 2020 Abstract: Background: We aimed to explore the relationship between hypoxia-inducible factors-1α (HIF-1α) and In-cRNA nuclear-enriched abundant transcript 1(NEAT1),and their functions on hepatocellular carcinoma (HCC) underhypoxia. Methods: HIF-1c and NEAT1 levels in HCC tissues and corresponding non-tumor tissues were determinedby qRT-PCR, and the correlations of their levels in HCC tissues were analyzed by Pearson test. The relationship be-tween overall survival and the two genes (HIF-1a and NEAT1) for HCC patients was detected by log-rank test. Clinico-pathological features of NEAT1 in HCC patients were collected. HIF-1a and NEAT1 levels in HCC cells were measuredby qRT-PCRand Western blot, and their relationship was determined by co-immunoprecipitation (Co-IP) assay. Cellviability, migration and invasion were detected by CCK-8, scratch wound healing and transwell assay, respectively.The interaction of NEAT1 with HIF-1a in tumor development was determined by xenograft tumor assays in nudemice. Results: NEAT1 and HIF-1a were highly expressed and showed a positive relationship in HCC tissues, andspecifically, higher NEAT1 expression was positively associated with advanced TNM stage and metastasis in HCC pa-tients. Up-regulated NEAT1 or HIF-1 in HCC patients had poorer prognosis. NEAT1 was induced by HIF-1a and sup-pressed by siHIF-1a. NEAT1 overexpression further promoted development of HCC under hypoxia while promotingcell viability, migration and invasion and suppressing apoptosis, and such effects were reversed by down-regulatingHIF-10. NEAT1 overexpression promoted tumor growth, which was reversed by down-regulating HIF-1a. Conclusion:HIF-1c knockdown inhibits NEAT1 expression, which suppresses progression of HCC and improves its prognosis. Keywords: HIF-1a, NEAT1, hepatocellular carcinoma, hypoxia, prognosis Hepatocellular carcinoma (HCC), which acco-unts for approximately 90% of primary livercancers, is the main type of primary livercancer and causes 700,000 deaths annuallyworldwide [1,2]. Meanwhile, HCC is also amajor cause of cancer-related deaths in China[3]. Though some therapeutic options, such assystemic chemotherapy, targeted therapy, sur-gical resection, and liver transplant, have beenadopted for the management of HCC [4], HCChas a high tolerance to the majority of anti-cancer treatments, and therefore the 5-yearsurvival rate of HCC patients is still low [5].Although efforts have been devoted to under-standing basic cellular events in HCC, the exact mechanisms of liver carcinogenesis still remainunclear [6]. Thus, it is highly necessary to dis-cover effective prognostic markers and poten-tial molecular mechanism involved in HCC car-cinogenesis so as to develop new targetedtherapeutic methods for improving the progno-sis of HCC patients. Hypoxia, which can be commonly found in gen-eral solid tumors, is often involved in poor prog-nosis. It promotes cancer cell invasion andmetastasis by activating related gene expres-sions via hypoxia-inducible factors (HIFs) [7].Hypoxia-inducible factor 1 (HIF1), a heterodi-meric transcription factor composed of an asubunit (HIF-1a) and a B subunit (HIF-1B), par-ticipates in hypoxic signalingpathways,and HIF- 1a is found to be the most mature factor instudying the relationship between HIF-1 andtumor [8]. HIF-1a controls cellular and systemichomeostatic responses to oxygen availability[9]. Under hypoxia, it recognizes and binds tohypoxia response elements (HREs), activatingthe transcription of numerous genes to modu-late pro-oncogenic events [10, 11]. Pathogene-sis of HCC is involved in tumor hypoxia and theactivation of HIF [12]. In HCC, HIF-1a exerts atumorigenic effect by promoting cell migrationand invasion [131. In recent years, multiple long non-coding RNAs(IncRNAs))have been reported to serve asimportant regulators in different cancers [14,15], and to be associated with the promotion orinhibition of typical cancer hallmarks, includingcontinuous proliferation, surpassing apoptosis,drug resistance, invasion, and metastasis [16].Structures and functions of IncRNA nuclear-enriched abundant transcript 1 (NEAT1) havebeen investigated widely for its significant rolein numerous cancer-related pathological chan-ges and metastasis [17].However, the role ofNEAT1 in HCC under hypoxia remains unclear. The current study explored the expressions ofHIF-1o and NEAT1, their correlations in HCC tis-sues and their relationships with prognosis forHCC patients. We also investigated the rela-tionship between HIF-1a and NEAT1 and theirroles in HCC progression in vitro under hypoxiaand normoxia, and in tumor progression invitro. Materials and methods Ethics statement All patients signed the written informed con-sent before surgery, and Research Ethics Com-mittee of Jiangsu Cancer Hospital (JCH2018-0504298) approved the current study accord-ing to the Declaration of Helsinki. In this study,all the animals were used in accordance withthe Guidelines of the China Council on AnimalCare and Use, and the experiments wereapproved by the Committee of ExperimentalAnimals of Jiangsu Cancer Hospital (JCH20-19060326). Efforts have been made to mini-mize pain or discomfort caused to the animals.The animal experiments were performed inJiangsu Cancer Hospital. Clinical specimens Ninety-eight patients who were diagnosed ashepatocellular carcinoma (HCC) and receivedroutine hepatic resection in Jiangsu CancerHospital from 02/16/2018 to 03/16/2019were included in this study. The patients had noradiotherapy or chemotherapy history and weretreated by surgery, and fresh tumor sampleswere obtained from their resected specimens.The clinicopathological characteristics (gender,age, tumor size, HBsAg, liver cirrhosis, histo-logical differentiation, TNM stage, and metas-tasis) are displayed in Table 1. Tumors andadjacent fresh non-tumor tissues were frozenimmediately in liquid nitrogen and stored at-80°C after the resection. Prognosis analysis Survival of HCC patients in relation to HIF-1αwas analyzed by Kaplan-Meier and log-ranktest based on data from The Cancer GenomeAtlas (TCGA). Overall survival of NEAT1 in HCCpatients was analyzed by Kaplan-Meier andlog-rank test based on data from follow-up time(60 months). Cell hypoxia culture Two human liver cancer cell lines (Hep3B andSK-Hep1) were purchased from American TypeCulture Collection and cultured in DMMEEM(12100, Solarbio, China) containing 10% fetalbovine serum (FBS;11011-8611,Solarbio). Forhypoxia treatment, the cells were culturedin an in vivo Hypoxia Work Station (RuskinnTechnology Ltd.) under normoxia (21%oxygen)or hypoxia (gas mixture of 1% oxygen, 5% CO,,and 94%N,). Cell transfection To construct plasmids that express NEAT1,the full-length human NEAT1 sequence wassynthesized by RiboBio and subcloned intothe pcDNA3.1 vector (V79020, ThermoFisher,USA). For HIF-1a knockdown, 100 pmol ofHIF-1a siRNA (siHIF-1a) (siRNA; GenePharma,Shanghai, China) and siRNA negative control(NC) were respectively transfected into thecells. The empty vector was considered as acontrol (pc-Control) of pcDNA-NEAT1. NC ser-ved as a control of siHIF-1a. After seeded into6-well plates, the cells were cultured for 24 h, Table 1. The relationship between NEAT1 and clinical characteristics of HCC patients Characteristics N Low NEAT1 expression I High NEAT1 expression P (n=45) (n=53) Gender Male 80 37 43 0.019 0.890 Female 18 8 10 Age (years) <55 55 26 29 0.093 0.761 ≥55 24 Tumor size (cm) <5 29 0.277 0.598 25 24 HBsAg Negative 5 5 0.075 0.785 Positive Liver cirrhosis Absence 0.002 0.960 Presence Histological differentiation Well 0.033 0.984 Moderate Poor TNM stage l + Il 5.699 0.017 II1+IV Metastasis No 7.642 0.006 Yes Table 2. Primer sequences used for quantita-tive reverse transcription-polymerase chainreaction (qRT-PCR) Genes Primer sequences (5'-3') HIF-1a Forward GAACGTCGAAAAGAAAAGTCTCG Reverse CCTTATCAAGATGCGAACTCACA NEAT1 Forward CAGTTAGTTTATCAGTTCTCCCATCCA Reverse GTTGTTGTCGTCA CCTTTCAACTCT GAPDH Forward GCTTCGGCAGCACATATACTAAAAT Reverse CGCTTCACGAATTTGCGTGTCAT and then transfected by Lipofectamine 2000(11668.,ThermoFisher.,USA)followingtheinstructions. The cells were collected aftertransfection for48 h. Quantitative reverse transcription-polymerasechain reaction (qRT-PCR) Total RNAs were isolated from the collected tis-sues and cells by Trizol reagent (15596018,Invitrogen). PrimeScriptTM RT Master Mix wasused for RT-PCR (RR036B, Takara). Quantita-tive PCR was performed in 7300 real-time PCRsystem (Applied Biosystems, USA) using TBGreen@ Premix Ex TaqTM ⅡI (RR820Q, Takara).The conditions were set as follows: incubationat 95°C for 30 min, followed by amplification at95°C for 5 sec and at 60°C for 34 sec for 40 cycles. Expressions of genes were normalizedto that of GAPDH (Sangon Biotech) using 2-AcTmethod [18].The sequences of primers usedare displayed in Table 2. Western blot Total proteins were extracted from the cellsusing RIPA buffer (R0010,Solarbio). The super-natant from the total proteins was collectedand the concentration was determined by BCAprotein assay kit (PC0020,Solarbio). The super-natant (30 pg/lane) was loaded for SDS gel andtransferred onto PVDF membranes (FFP32,Beyotime, China) by using an electroblottingapparatus. Non-specific bindings were blockedby incubating the membranes in 5%6 (w/v)skimmed milk in Tris-buffered saline containing0.5% Tween-20 (w/v) buffer at 37°C for 1 h.The membranes were probed by specific pri-mary antibodies [anti-HIF-1α (Rabbit, 1:1000,ab51608, Abcam, USA) and anti-GAPDH anti-body (Mouse, 1:500, ab8245, Abcam, USA)] at4°C overnight, and washed by TBS containing0.1% Tween-20. The membranes were thenincubated with HRP-conjugated secondary anti-bodies (Goat Anti-Mouse, 1:2000, ab205719,Abcam, USA; Goat Anti-Rabbit,1:2000, ab-205718, Abcam, USA) at 1:1000 dilution at37°C for 1 h. After washing the membranes 3times at an interval of 10 min, the signals were visualized by ECL detection kit (ECL, Premega,USA) and normalized to GAPDH. Co-immunoprecipitation (Co-IP) assay Co-IP assay was performed as described previ-ously [19]. Briefly, the total proteins wereextracted from the cell lines and the concentra-tion was determined. The total proteins wereincubated with anti-HIF-1α (ab51608, Abcam),or with control IgG antibodies and protein-Gagarose beads (Beyotime) overnight at 4°C.The immunoprecipitates were boiled and thenanalyzed by Western blot. Cell counting kit-8 (CCK-8) assay Transfected cells (5x10° per well) were seededin 96-well plates and cultured for 24 h. Then,CCK-8 solution (10 pL/well, CK04, Japan) wasadded into the plates for further incubation for2 h. The absorbance was measured at 450 nmusing a microplate reader (SpectraMax iD5,Molecular Devices, US). Scratch wound healing assay The cells were cultured in 6-well plates to reach80-90% confluence, scratched by a 10 pl tipand further cultured for 24 h. The migrationrate was calculated from photomicrographsunder a 100x inverted microscope (Ts2r-FL,Nikon,Japan). Invasion assays The cells were cultured in Transwell cham-bers (8 mm pores, Corning Inc., Corning, USA)composed of Transwell@-precoated MatrigelTMmembrane filter inserts. The 10% FBS wasadded into the lower chamber and served asthe chemoattractant. After 24 h, the remainingcells were gently removed by cotton swabs,while those adhered to lower surface were fixedby 4% methanol for 30 min and stained by 0.1%crystal violet solution for 20 min at 37°C andphotographed. Flow cytometryassay Cell apoptosis after treatment was determinedby flow cytometry using an Annexin V-FITCApoptosis Detection Kit (CA1020, Solarbio,China). Briefly,the cells were treated and cul-tured in 6-well plates. After washed by coldsterile PBS, the cells were counted, resuspend- ed in 1X binding buffer, and incubated with 5 pLof Annexin V-FITC and blended gently for 10min, followed by incubation with 5 pL of prop-idium iodide (PI) for 5 min in the dark. Then thecells were diluted with PBS to 500 pL. Afterthat, the apoptotic rate was evaluated by flowcytometry in flow cytometer AccuriTM C6 (BDBiosciences) and the data were analyzed byCell Quest software 3.3 (Becton-Dickinson). Animal experiments in vivo For tumorigenicity in vivo, 20 male BALB/c athy-mic nude mice (aged 6 weeks old) were ran-domly divided into four groups, with five mice ineach group. SiHIF-1a, pc-NEAT1-transfected orSiHIF-1a, and pc-NEAT1 co-transfected Hep3Bcells (1x10" cells in 100 pL) were injected sub-cutaneously into the left flanks of the nudemice. In the control group, tumor growth wasmeasured every week and when the diameterreached 1 cm, the mice were sacrificed andtumors were weighted. The tumor volume wascalculated according to the followingequation:V=0.5x D×d² (V=volume; D = longitudinaldiameter and d = latitudinal diameter). The tis-sue samples were paraffin-embedded and sec-tioned, and the expressions of HIF-1a proteinand NEAT1 were determined by immunohisto-chemistry and qRT-PCR. Immunohistochemistry Immunohistochemistry was performed as pre-viously described [6]. The tissues were sec-tioned into 4 pm thick, treated with antigenretrieval by heating ethylene diamine tetra ace-tic acid buffer in a microwave oven. Endogenousperoxidase activity was blocked by incubatingthe sections in 3% H,O, for 30 min and thenrinsed in PBS three times at an interval of 3min. The sections were incubated with mousemonoclonal antibody against HIF-1α (ab1, 1:500, Abcam) at 4°C overnight and then incu-bated with secondary antibody (Goat Anti-Mouse, 1:2000, Abcam). The DAB HorseradishPeroxidase Color Development Kit (PO203,Beyotime) was used to develop color for target-ing proteins. Next, the sections were redyed byHematoxylin Staining Solution (C0107, Beyo-time). Statistical analysis The data were shown as mean ± standard devi-ation (S.D.). Chi-squaretest was used to deter- expression levels group E · low higl (low.i +1(high,1) Figure 1. Correlations and relative expressions of HIF-1a and NEAT1 in HCC tissues and their relationships withoverall survival of HCC patients. (A and B) HIF-1α (A) and NEAT1 (B) expressions were detected by qRT-PCR and nor-malized to GAPDH expression in 98 pairs of HCC tissues compared with adjacent nontumourous liver specimens.(C) Spearman's rank correlation analysis was performed to analyze the correlations of HIF-1a and NEAT1 in HCCtissues. (D and E) Kaplan-Meier survival curve and log-rank test were used to evaluate whether HIF-1a and NEAT1expression levels were associated with overall survival rate. ***P <0.001 vs. Adjacent tissue. The data were shownas mean ± standard deviation (S.D.). Abbreviations: HIF-1a,hypoxia-inducible factors-1o; NEAT1, IncRNA nuclear-enriched abundant transcript 1; qRT-PCR, quantitative reverse transcription-polymerase chain reaction; HCC, hepa-tocellular carcinoma. mine the correlation between NEAT1 and clini-cal features in liver cancer patients. Pairedt-test was performed for paired samples.Correlation coefficients (r) were analyzed byPearson correlation. Kaplan-Meier plots wereanalyzed by log-rank test. Comparisons amongmultiple groups were conducted by one-wayANOVA followed by Bonferroni-t post hoc test.Statistical analyses were performed usingSPSS 17.0 software (SPSS, Inc., Chicago, IL,USA), and P<0.05 was considered to be of sta-tistical significance. Results NEAT1 and HIF-1a were differentially overex-pressed in HCC tissues and their overexpres-sions were associated with poorer prognosis ORT-PCR results showed that NEAT1 and HIF-1a were obviously increased in HCC tissuescompared with those in adjacent normal tis-sues (Figure 1A and 1B, P<0.001). Pearson correlation analysis revealed that HIF-1a ex-pression was positively correlated with NEAT1expression in HCC specimens (Figure 1C, P <0.05). Data on differential expression of HIF-1a inHCC cancer were collected from TCGA. The pro-portion of surviving HCC patients in the HIF-1ahigh group (n=184) was significantly lower thanthat in the HIF-1a low group (n=185), suggest-ing that HCC patients with low HIF-1a expres-sion could have a better prognosis (Figure 1D,P=0.0026). As shown in Table 1, HCC patients were dividedinto two groups, namely, low NEAT1-expressiongroup(n=45) and high NEAT1-expression group(n=53), according to the cut-off value, whichwas defined as the median value of the NEAT1level. Clinicopathological analysis showed thatthere were no statistically significant differenc-es in gender, age, tumor size, HBsAg,and histo-logical differentiation liver cirrhosis between Figure 2. NEAT1 was induced by HIF-1a and HIF-1a interacted with the hypoxia response element (HRE) in thepromoter region of NEAT1. A-F. The mRNA and protein levels of HIF-1a were detected in Normal (non-transfectedcontrol), Normal+ NC (siRNA non-targeting control), Normal+ siHIF-1α (siRNA-HIF-1α), Hypoxia, Hypoxia + NC,Hypoxia+siHIF-1o groups in Hep3 and SK-Hep1 cells by qRT-PCR and Western blot. G. The mRNA levels of NEAT1were detected in Normal, Normal + NC, Normal +siHIF-1a, Hypoxia, Hypoxia +NC, Hypoxia + siHIF-1a groups inHep3 cells by qRT-PCR. H. The mRNA levels of NEAT1 were detected in Normal, Normal + NC, Normal + siHIF-1a,Hypoxia, Hypoxia + NC, Hypoxia +siHIF-1o groups in SK-Hep1 cells by qRT-PCR. I. ChIP assays were performed todetect whether HIF-1a directly bound to the HRE of NEAT1 promoter. GAPDH served as an internal control. *P <0.05 or **p<0.01 or ***P <0.001 vs. Normal+ NC. ###p<0.001 vs. Normal. ^^^P<0.001 vs. Hypoxia + NC. Thedata were shown as mean ± standard deviation (S.D.). Abbreviations: HIF-1a, hypoxia-inducible factors-1o; NEAT1,IncRNA nuclear-enriched abundant transcript 1; qRT-PCR, quantitative reverse transcription-polymerase chain reac-tion; NC, negative control. the low NEAT1 and high NEAT1 groups (Table1,P > 0.05); However, a high level of NEAT1was found to be significantly associated withadvanced tumor-node-metastasis (TNM) stage(Table 1,P=0.017) and metastasis of HCC(Table 1, P=0.006). Kaplan-Meier plots showed that HCC patientsin the high NEAT1-expression group (n=53) hadlower overall survival rate (15%-25%) compared with the low NEAT1-expression group (n=45)(Figure 1E, P=0.018). HIF-1a silencing reduced NEAT1 level inHep3B and SK-Hep1 cells under hypoxia SiHIF-1a significantly reduced the protein andmRNA levels of HIF-1a in Hep3B and SK-Hep1cells (Figure 2A-F, P <0.05 or P < 0.01 or P <0.001). Nevertheless, the protein and mRNA levels of HIF-1a were highly expressed in Hep3Band SK-Hep1 cells under hypoxia (Figure 2A-F,P<0.001), which, however, could be partiallyreversed by silencing HIF-1 (Figure 2A-F, P <0.001). Under hypoxia, NEAT1 was highly expressed inHep3B (Figure 2G, P< 0.001) and SK-Hep1cells (Figure 2H, P < 0.001), while this effectwaspartially reversed1 by HIF-1o silencing(Figure 2G and 2H, P <0.001). In addition, wefound that HIF-1a bound to hypoxia responseelements (HRE) in gene promoters of NEAT1 inhypoxic Hep3B and SK-Hep1 cells (Figure 2l),suggesting that HIF-1a can regulate protein-coding genes of NEAT1. HIF-1a silencing suppressed the developmentof HCC by negatively regulating NEAT1 level inHep3B and SK-Hep1 cells under hypoxia Hep3B and SK-Hep1 cells in the hypoxia, pc-control, pc-NEAT1, pc-NEAT1 +siHIF-1a, siHIF-1a groups were exposed to hypoxia, and weobserved that hypoxia or NEAT1 overexpres-sion significantly increased the level of NEAT1in Hep3B (Figur 3A, P<0.001) and SK-Hep1cells (Figure 3B, P<0.001), while HIF-1a silenc-ing greatly reduced the protein level of NEAT1(Figure 3A and 3B, P<0.001). NEAT1 overexpression further promoted theviability of Hep3B (Figure 3C, P <0.001) andSK-Hep1 cells (Figure 3D, P <0.01) under hy-poxia. However, HIF-1a silencing significantlysuppressed the cell viability under hypoxia(Figure 3C and 3D, P <0.01), which was par-tially reversed NEAT1 overexpression (Figure3C and 3D, P<0.001). Moreover, the apopto-sis rates of Hep3B (Figure 3E, P<0.001) andSK-Hep1 cells (Figure 3F, P <0.001) wereslightly increased under hypoxia. HIF-1a silenc-ing significantly increased the apoptosis ratesof Hep3B (Figure 3E, P <0.001) and SK-Hep1cells (Figure 3F, P < 0.001) under hypoxia,which was partially reversed by NEAT1 overex-pression (Figure 3E and 3F, P<0.001). As shown in Figure 4A-E, NEAT1 overexpres-sion further promoted migration and invasionunder hypoxia (P <0.001), while HIF-1a silenc-ing noticeably suppressed migration and inva-sion under hypoxia (P<0.001), and the effectsof HIF-1a silencing were partially reversed byNEAT1 overexpression (P <0.001). Knocking down HIF-1a inhibited tumor growthin vivo through negatively regulating NEAT1expression To evaluate the effects of siHIF-1c and NEAT1on the tumor growth of HCC cells in vivo, weestablished a xenograft model in which theHep3B cells treated with siHIF-1a, or pc-NEAT1,or siHIF-1a and pc-NEAT1 were subcutaneouslyinjected into the fland of athymic mice, respec-tively. The xenograft model was allowed todevelop measurable tumors. The volume (Fig-ure 5A and 5C, P <0.001) and weight (Figure5A and 5D, P <0.001) of the tumors in thesiHIF-1a group were significantly smaller andlighter than those in the control group, which,however, were reversed by pc-NEAT1 (Figure5A, 5C and 5D, P <0.001). Next, qRT-PCR analysis of NEAT1 expressionand immunostaining analysis of HIF-1a proteinexpression were performed in resected tumortissues. As described in Figure 5E, the relativelevel of NEAT1 expression in the siHIF-1agroupwas significantly lower than that in the controlgroup(P <0.001), which was reversed by pc-NEAT1 (P<0.001). As described in Figure 5B,siHIF-1a reduced the protein level of HIF-1a,while pc-NEAT1 did not affect the protein levelof HIF-1α. Collectively, our in vivo and in vitro data sup-ported that NEAT1 can act as an oncogenicIncRNA in HCC, and its expression can be sup-pressed by HIF-1o knockdown. Discussion In the study, the data showed that NEAT1 andHIF-1a were highly expressed in HCC. Consis-tent with our findings, a recent study revealedthat NEAT1 was obviously up-regulated in HCCtissues and cells [20]. We also found that high-er expressions of HIF1a and NEAT1 were asso-ciated with poorer prognosis in HCC patients.Moreover, studies showed that in human HCCtissues,HIF1x is highly elevated and is associ-ated with poorer prognosis [21, 22]. Li G et al.reported that IncRNA plays an important rolein tumorigenesis, subsequent prognosis andmetastasis of HCC [23], which is consistentwith our findings that higher NEAT1 expressionwas closely associated with advanced TNMstage, metastasis and prognosis in HCC pati-ents. Liu Z et al. suggested that in HCC tissues The role of HIF-1a and NEAT1 in hepatocellular carcinoma Figure 3. HIF-1a silencing suppressed the development of HCC by negatively regulating NEAT1 level in Hep3B andSK-Hep1 cells under hypoxia. (A and B) The mRNA levels of NEAT1 were detected in Normal (non-transfected andnon-hypoxia control), Hypoxia, pc-Control (non-targeting and hypoxia control), pc-NEAT1 (NEAT1 transfected andhypoxia), pC-NEAT1+siHIF-1α (NEAT1 and siRNA-HIF-1α co-transfected under hypoxia), siHIF-1a groups in Hep3 (A)and SK-Hep1 cells (B) by qRT-PCR. (C and D) The viability rates were detected in Normal, Hypoxia, pc-Control, pc-NEAT1, pc-NEAT1+siHIF-1a, siHIF-1o groups in Hep3 (C) and SK-Hep1 cells (D) by CCK-8. (E and F) The apoptosisrates were detected in Normal, Hypoxia, pc-Control, pc-NEAT1, pc-NEAT1+ siHIF-1a, siHIF-1a groups in Hep3 (C)and SK-Hep1 cells (D) by flow cytometry assay.GAPDH served as an internal control. *P <0.05 or ***p<0.001vs.Normal. ###P<0.001 vs. pc-Control. ^^^P<0.001vs. pc-NEAT1.AAAP<0.001 vs. pc-NEAT1+siHIF-1a. The data wereshown as mean ± standard deviation (S.D.). Abbreviations: HIF-1a, hypoxia-inducible factors-1o; NEAT1, IncRNAnuclear-enriched abundant transcript 1; qRT-PCR, quantitative reverse transcription-polymerase chain reaction;CCK-8, cell counting kit-8. NEAT1 overexpression is an independent riskfactor associated with the prognosis of HCCpatients [24]. Besides, our data directly con-firmed that HIF-1a expression was positivelycorrelated with NEAT1 expression in HCC tis-sues. Taken together, expressions of HIF-1aand NEAT1 have high predictive and diagnosticvalues and can serve as prognostic markersand treatment targets for HCC. Hypoxia is a general symptom in solid cancersand is particularly common in HCC due to itsrapid growth [25]. Hypoxia is closely related totumor progression and negatively affects over-all prognosis [26]. Our results demonstratedthat HIF-1a and NEAT1 were highly expressedin HCC cells under hypoxia, suggesting thathypoxia caused poor prognosis to HCC patientspossibly through overexpressing HIF-1a andNEAT1. Moreover, Co-IP results demonstratedthat HIF-1a promotedNEAT1transcriptionthrough directly binding to the HRE in the pro-moter region of NEAT1, which were consistentwith the correlation of HIF-1c and NEAT1 in HCCtissues and the result that NEAT1 expressionwas inhibited by down-regulating the expres-sion of HIF-1a. In view of this, we suggestedthat the overexpression of NEAT1 was inducedpartly by HIF-1a directly binding to the NEAT1promoter in HCC cells under hypoxia.Therefore,we elucidated that it might be a promising ther-apeutic option of HCC to inhibit the expressionof NEAT1 by HIF-1a silencing. In the present study, the results demonstratedthat NEAT1 further promoted the HCC celldevelopment, evidenced by increased cell via-bility, migration and invasion, and suppressedapoptosis in HCC cells under hypoxia, while HIF-1a silencing further promoted the above phe-nomena. Consistently, previous study showedthat down-regulating the expression of NEAT1not only suppressed proliferation, migration, and invasion of HCC cells, but also promotedapoptosis of HCC cells [27]. We also found thatthe effects of NEAT1 overexpression in HCCcells under hypoxia were partially reversed byHIF-1a silencing. Migration refers to any direct-ed cell movement within the body,and invasionof carcinomas is the penetration of tissuebarriers [28]. The migration and invasion abili-ties of cancer cells enables diseases to spreadthrough changing position within tissues orfrom the initial tumor [29]. Cancer cell migra-tion and invasion into surrounding tissues andvasculature are the pivotal and initial events incancer metastasis [29]. Frequent tumor metas-tasis will bring dismal outcome to HCC patients[30].Thus, inhibition of HIF-1a might suppressHCC development and improve prognosis bynegatively regulating the expression of NEAT1under hypoxia. In addition, the results fromxenograft tumor assays in nude mice furtherrevealed that HIF-1a silencing suppressed HCCmalignant progression in vitro through inhibit-ing the expression of NEAT1. In brief, our results provide novel findings thatimprove the current understanding on HCCpathogenesis and provide a promising strate-gy for the diagnosis and treatment of HCC.However, HCC progression is a complex pro-cess involving the most gene networks andchanges in signaling pathways, many of whichstill need to be illuminated [31]. For example,Ling Z A et al. [27] reported that NEAT1 pro-motes the development of HCC through inter-acting with several tumor-related genes suchas SP1 and KAT2A; Mang Y et al. [32] foundthat NEAT1 promotes cell proliferation andinvasion through modulating hnRNP A2 expres-sion in HCC cells. These findings suggestedthat the mechanism of HIF-1c and NEAT1 inHCC under hypoxia may involve other down-stream targets, yet more experiments are The role of HIF-1a and NEAT1 in hepatocellular carcinoma Figure 4. HIF-1o silencing suppressed the development of HCC by negatively regulating NEAT1 level in Hep3B andSK-Hep1 cells under hypoxia. (A, B) Migration rates were detected in Normal (non-transfected and non-hypoxia con-trol), Hypoxia, pc-Control (non-targeting and hypoxia control), pc-NEAT1 (NEAT1 transfected and hypoxia), pc-NEAT1+ siHIF-1α (NEAT1 and siRNA-HIF-1α co-transfected under hypoxia), siHIF-1a groups in Hep3 (A and B) and SK-Hep1cells (C and D) by scratch wound test.(E-H) Invasion rates were detected in Normal, Hypoxia, pc-Control, pc-NEAT1,pc-NEAT1+ siHIF-1a, siHIF-1a groups in Hep3 (E and F) and SK-Hep1 cells (G and H) by transwell assay. ***p <0.001 vs. Normal.###P<0.001 vs. pc-Control. ^^^P<0.001 vs. pc-NEAT1.A^AP<0.001 vs. pc-NEAT1+siHIF-1a. Thedata were shown as mean ± standard deviation (S.D.). Abbreviations: HIF-1a, hypoxia-inducible factors-1o; NEAT1,IncRNA nuclear-enriched abundant transcript 1. Figure 5. Knockdown of HIF-1o inhibited tumor growth in vivo through negatively regulating NEAT1 expression.A.Photographs of representative tumor diameter of nude mice. B. Immunohistochemistry assays were performedto detect HIF-1a protein expression in tumor tissues of Control (no injection of Hep3B cells), siHIF-1a (injection ofHep3B cells transfected with siRNA HIF-1α), siHIF-1α+ pc-NEAT1 (injection of Hep3B cells transfected with siRNAHIF-1a and NEAT1) and pc-NEAT1 (injection of Hep3B cells transfected with NEAT1) groups. C. Tumor volumes weremeasured in tumor tissues of siHIF-1a, siHIF-1α+ pc-NEAT1 and pc-NEAT1 groups. D. Tumor weights were mea-sured in tumor tissues of siHIF-1a, siHIF-1α+pc-NEAT1 and pc-NEAT1 groups. E. The mRNA levels of NEAT1 weredetermined in tumor tissues of siHIF-1a, siHIF-1α+ pc-NEAT1 and pc-NEAT1 groups by qRT-PCR. GAPDH served asan internal control. ***P <0.001 vs. Control. ###P<0.001 vs. siHIF-1a. ^^^P<0.001 vs. siHIF-1α+pc-NEAT1. Thedata were shown as mean ± standard deviation (S.D.). Abbreviations: HIF-1a, hypoxia-inducible factors-1o; NEAT1,IncRNA nuclear-enriched abundant transcript 1; qRT-PCR, quantitative reverse transcription-polymerase chain reac-tion. required to be performed in appropriate cellsor animal models to confirm this hypothesis.However, the generate more data of somemechanistic studies should be further explorefor supporting the observation and conclusionin this study. Taken together, the study demonstrates thatNEAT1 acts as an oncogene by promotingmalignant development, and that higher ex-pression of NEAT1 predicts a poorer prognosis of human HCC. The expression of NEAT1 couldbe suppressed by down-regulating the expres-sion of HIF-1α. Acknowledgements This work was supported by the Young TalentsProgram of Jiangsu Cancer Hospital [GrantNumber QL201801]; the NationalNaturalScience Foundation of China [Grant Number81872485]. Thanks for the financial supports. Disclosure of conflict of interest None. Address correspondence to: Wenrong Shen, De-partment of Radiology, Jiangsu Cancer Hospital,Jiangsu Institute of Cancer Research, The AffiliatedCancer Hospital of Nanjing Medical University, No.42, Baiziting, Xuanwu District, Nanjing210000,Jiangsu, China. Tel: +86-025-83283502; E-mail:shenwr_wrong@163.com References ( [1] Rawat D, Shrivastava S, Naik RA, Chhonker SK, Mehrotra A and K o iri RK. An overview o f natu- ral plant products i n the treatment of hepato- cellular carcinoma. Anticancer Agents Med Chem 2018;18:1838-1859. ) ( [21 Pollicino T and S aitta C . O ccult hepatitis B virus and hepatocellular carcinoma. World J Gastroenterol 2014;37:439-440. ) [3] Zhu ZX, Huang JW, Liao MH and Zeng Y. Treat-ment strategy for hepatocellular carcinoma inChina: radiofrequency ablation versus liver re-section. JpnJ Clin Oncol 2016;46:1075-1080. [4] Shiani A, Narayanan S, Pena L and FriedmanM. The role of diagnosis and treatment of un-derlying liver disease for the prognosis of pri-mary liver cancer. Cancer Control 2017;24:1073274817729240. [5] Siegel RL, Miller KD and Jemal A. Cancer sta-tistics, 2017. CA Cancer J Clin 2017;67: 7-30. [6] Wang F, Ying HQ, He BS, Pan YQ, Deng QW, SunHL, Chen J, Liu X and Wang SK. UpregulatedIncRNA-UCA1 contributes to progression of he-patocellular carcinoma through inhibition ofmiR-216b and activation of FGFR1/ERK sig-naling pathway. Oncotarget 2015; 6:7899-7917. ( [71 L u X a nd Kang Y. H y poxia and hy p oxia-induc-ible factors: master regulators o f m e tastasis. Clin Cancer Res 2010; 16:5928-5935. ) [8] Fallone F, Britton S, Nieto L, Salles B andMuller C. ATR controls cellular adaptation tohypoxia through positive regulation of hypoxia- ( i nducible f actor 1 ( HIF-1) expression. O nco- gene 2013;32: 4387-4396. ) ( 9] S nell CE, Turley H, Mclntyre A, Li D, Masiero M,Schofield C J, Gatter KC, Harris AL and PezzellaF. Proline-hydroxylated hypoxia-inducible factor1alpha(HIF-1alpha) upregulation in human tu- mours. PLoS One 2014;9: e88955. ) ( [10] ]L L iu L, Zhao X , Zou H, B ai R , Yang K and Tian Z . Hypoxia Promotes gastric cancer malignancy partly through the HIF-1alpha dependent tran-scriptional activation of the long non-coding RNA GAPLINC. Front Physiol 2016; 7 :420. ) [111Nagaraju GP, Park W, Wen J, Mahaseth H,Landry J, Farris AB, Willingham F, Sullivan PS,Proia DA, El-Hariryl, Taliaferro-Smith L, Diaz Rand El-Rayes BF. Antiangiogenic effects ofganetespib in colorectal cancer mediatedthrough inhibition of HIF-1alpha and STAT-3.Angiogenesis 2013; 16:903-917. ( [12] C (hen C a n d Lou T. Hypoxia inducible factors inhepatocellular carcinoma. Oncotarget 2017;8: 46691-46703. ) ( [13] ]F F eng W, X u e T, Huang S, Shi Q, Ta n g C, Cu i G, Yang G, Gong H and G u o H. HIF-1alpha p r o- motes the migration and i nvasion of hepato-cellular carcinoma cells via the IL-8-NF-kappaB axis. Cell Mol Biol Lett 2018;23:26. ) ( [14] Li D, He J, Qian X, Xu P, Wang X, Li Z, Qian J and Yao J. The involvement of IncRNAs in the devel- opment and progression of pancreatic cancer. Cancer Biol Ther 2017;18: 9 27-936. ) ( [15] H uang X, Zhi X, Gao Y, Ta N, Jiang H and ZhengJ. LncRNAs in p ancreatic c ancer. O ncotarget 2016;7:57379-57390. ) ( [161 Renganathan A and Fe l ley-Bosco E. L o ng non- coding RNAs in cancer and t h erapeutic poten-tial. Adv Exp Med Biol 2017 ; 1008:199-222. ) ( [17] F ang L, S un J, P an Z, Song Y, Z hong L, ZhangY,Liu Y, Zheng X and H uang P . Long n o n-coding RNA NEAT1 promotes he p atocellular carcino- ma cell p roliferation through t he regulation ofmiR-129-5p-VCP-IkappaB. AmJ Physiol Gastro- intest Liver Physiol 2017;313:G150-G156. ) ( [18] Livak K J and S c hmittgen TD. Analysis of rela- tive gene e xpression u s ing d i fferent real-timequantitative PCR and 2-AACT m ethod. Acta Ag- ronomica Sinica 2001;25:402-408. ) ( [19] L i Y, Gao Y, Cui T, Yang T, Liu L, Li T and Chen J.Retinoic acid facilitates toll-like r eceptor 4 ex - pression t o improve intestinal barrier function through r e tinoic acid r e ceptor be t a. Ce l l Physi-ol Biochem 2017;42:1390-1406. ) ( [20] W \ ang Z, Zou Q, Song M and Chen J. NEAT1 pro-motes cell p roliferation and i nvasion in hepato- cellular carcinoma by negative regulating miR- 613 expression. Biomed Pharmacother 2017; 94:612-618. ) ( [21] W ang M, Zh a o X, Zhu D, L iu T, L iang X, Liu F, Zhang Y, Dong X a nd Sun B. HIF-1alpha pro-moted vasculogenic m i micry formation in he- ) patocellular carcinoma through LOXL2 up-reg-ulation in hypoxic tumor microenvironment. JExp Clin Cancer Res 2017; 36: 60. [22]Yang SL, Liu LP, Jiang JX, Xiong ZF, He QJ andWu C. The correlation of expression levels ofHIF-1alpha and HIF-2alpha in hepatocellularcarcinoma with capsular invasion, portal veintumor thrombi and patients’clinical outcome.Jpn J Clin Oncol 2014; 44: 159-167. [23]Li G, Zhang H, Wan X, Yang X, Zhu C, Wang A,He L, Miao R, Chen S and Zhao H. Longnoncoding RNA plays a key role in metastasisand prognosis of hepatocellular carcinoma.Biomed Res Int 2014;2014:780521. [241Liu Z, Chang Q, Yang F, Liu B, Yao HW, Bai ZG,Pu CS, Ma XM, Yang Y, Wang TT, Guo W, ZhouXN and Zhang ZT. Long non-coding RNA NEAT1overexpression is associated with unfavorableprognosis in patients with hepatocellular carci-noma after hepatectomy: a Chinese popula-tion-based study. Eur J Surg Oncol 2017; 43:1697-1703. [25]Keith B, Johnson RS and Simon MC. HIF1alphaand HIF2alpha: sibling rivalry in hypoxic tu-mour growth and progression. Nat Rev Cancer2011;12:9-22. [26]Wu W, Hu Q, Nie E, Yu T, Wu Y, Zhi T, Jiang K,Shen F, Wang Y, Zhang J and You Y. Hypoxiainduces H19 expression through direct and in-direct Hif-1alpha activity, promoting oncogeniceffects in glioblastoma. Sci Rep 2017; 7:45029. [27]11Ling ZA, Xiong DD, Meng RM, Cen JM, Zhao N,Chen G, Li RL and Dang YW. LncRNA NEAT1promotes deterioration of hepatocellular carci-noma based on in vitro experiments, data min-ing, and RT-qPCR analysis. Cell Physiol Bio-chem2018;48:540-555. ( [28] K ramer N , Walzl A, U nger C, R osner M, Krupit-za G , Hengstschlager M and Dol z nig H. In v itrocell migration and i nvasion a ssays. M utat R e s 2013:752:10-24. ) [29]Duff D and Long A. Roles for RACK1 in cancercell migration and invasion. Cell Signal 2017;35:250-255. [30]Hua L, Wang CY, Yao KH, Chen JT, Zhang JJ andMa WL. High expression of long non-codingRNA ANRIL is associated with poor prognosisin hepatocellular carcinoma. Int J Clin ExpPathol 2015;8:3076-3082. ( [311Zucman-Rossi J, Villanueva A, N ault JC and LI-ovet JM. Genetic landscape and biomarkers ofhepatocellular carcinoma. Gastroenterology2015;149:1226-1239.e1224. ) ( [321 M ang Y, Li L, R an J, Zhang S, Liu J, Li L, Chen Y,Liu J, Gao Y and Ren G. Long noncoding RNANEAT1 promotes cell proliferation and invasion by regulating hnRNP A2 e xpression in hepato- cellular carcinoma c ells. Onco T argets T her 2017;10:1003-1016. ) m J Transl Res : 文章题目:Silencing of HIF-1α inhibited the expression of lncRNA NEAT1 to suppress development of hepatocellular carcinoma under hypoxia沉默HIF-1α可以抑制lncRNA NEAT1 的表达,从而抑制缺氧条件下肝细胞癌的发展文章出处:EAm J Transl Res 2020;12(7):3871-3883.中国南京医科大学附属肿瘤医院江苏省肿瘤医院放射科工作站使用情况:Ruskinn Hypoxia Work Station使用气体浓度:低氧(1% O2)摘要:本研究旨在探讨缺氧诱导因子-1α (HIF-1α)和lncRNA 核富集丰富转录本1 (NEAT1)之间的关系,以及它们在缺氧条件下肝细胞癌(HCC)中的作用。NEAT1 和HIF-1α在肝细胞癌组织中呈高表达并呈正相关,其中NEAT1 高表达与肿瘤淋巴结转移(TNM)晚期及远处转移呈正相关; NEAT1 或HIF-1α上调的HCC 患者预后较差。NEAT1由HIF-1α诱导,siHIF-1α抑制NEAT1 表达;NEAT1 的过表达进一步促进了缺氧条件下肝癌的发展,同时促进细胞活力、迁移和侵袭,抑制细胞凋亡,下调HIF-1α可逆转这种作用。NEAT1 过表达促进肿瘤生长,下调HIF-1α可逆转NEAT1 促进肿瘤生长的发生, 揭示下调HIF-1α可抑制NEAT1 的表达,抑制HCC 的进展,改善预后。Figure 1. Correlations and relative expressions of HIF-1α and NEAT1 in HCC tissues and their relationships with overall survival of HCC patients. (A and B) HIF-1α (A) and NEAT1 (B) expressions were detected by qRT-PCR and normalized to GAPDH expression in 98 pairs of HCC tissues compared with adjacent nontumourous liver specimens. (C) Spearman’s rank correlation analysis was performed to analyze the correlations of HIF-1α and NEAT1 in HCC tissues. (D and E) Kaplan-Meier survival curve and log-rank test were used to evaluate whether HIF-1α and NEAT1 expression levels were associated with overall survival rate. ***P < 0.001 vs. Adjacent tissue. The data were shown as mean ± standard deviation (S.D.). Abbreviations: HIF-1α, hypoxia-inducible factors-1α; NEAT1, lncRNA nuclear enriched abundant transcript 1; qRT-PCR, quantitative reverse transcription-polymerase chain reaction; HCC, hepa tocellular carcinoma.Figure 4. HIF-1α silencing suppressed the development of HCC by negatively regulating NEAT1 level in Hep3B and SK-Hep1 cells under hypoxia. (A, B) Migration rates were detected in Normal (non-transfected and non-hypoxia control), Hypoxia, pc-Control (non-targeting and hypoxia control), pc-NEAT1 (NEAT1 transfected and hypoxia), pc-NEAT1 + siHIF-1α (NEAT1 and siRNA-HIF-1α co-transfected under hypoxia), siHIF-1α groups in Hep3 (A and B) and SK-Hep1 cells (C and D) by scratch wound test. (E-H) Invasion rates were detected in Normal, Hypoxia, pc-Control, pc-NEAT1, pc-NEAT1 + siHIF-1α, siHIF-1α groups in Hep3 (E and F) and SK-Hep1 cells (G and H) by transwell assay. ***P < 0.001 vs. Normal. ###P < 0.001 vs. pc-Control. ^^^P < 0.001 vs. pc-NEAT1. ΔΔΔP < 0.001 vs. pc- NEAT1 + siHIF-1α. The data were shown as mean ± standard deviation (S.D.). Abbreviations: HIF- 1α, hypoxia-inducible factors-1α; NEAT1, lncRNA nuclear-enriched abundant transcript 1. HCC 组织中NEAT1 和HIF1α明显升高(图1A 和1B);相关性分析显示HIF-1α的表达与NEAT1 的表达呈正相关(图1C),HIF-1α高表达组存活的HCC 患者比例明显低于HIF-1α低表达组((图1D),提示HIF-1α低表达的HCC 患者预后较好; NEAT1 过表达进一步促进了缺氧条件下的迁移和侵袭,沉默HIF-1α显著抑制了缺氧条件下的迁移和侵袭,而NEAT1 过表达部分逆转了沉默HIF-1α的作用(图4A-H);表明HIF-1α沉默通过负向调节缺氧条件下Hep3B 和SK-Hep1 细胞NEAT1 水平抑制肝癌的发生发展。
确定


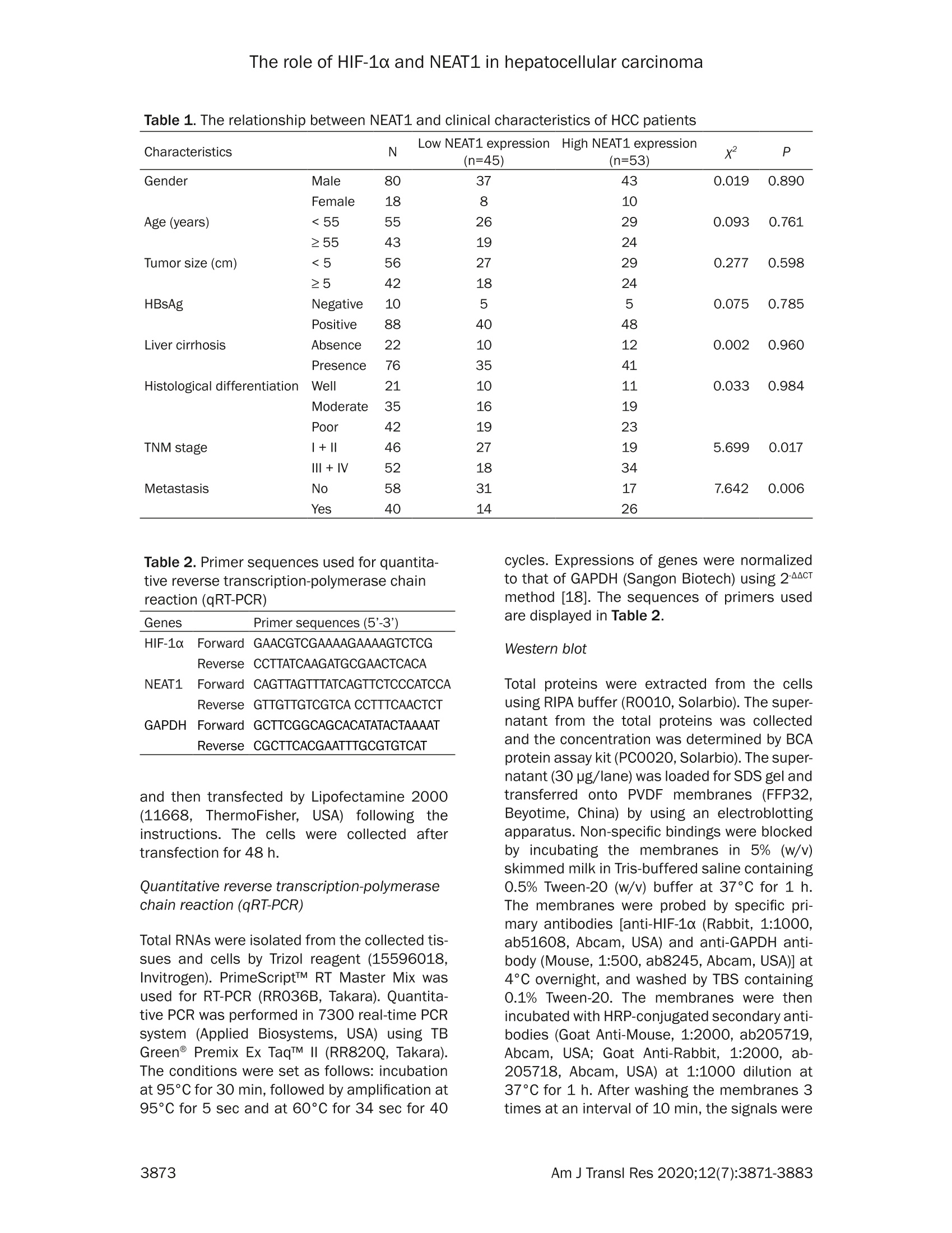

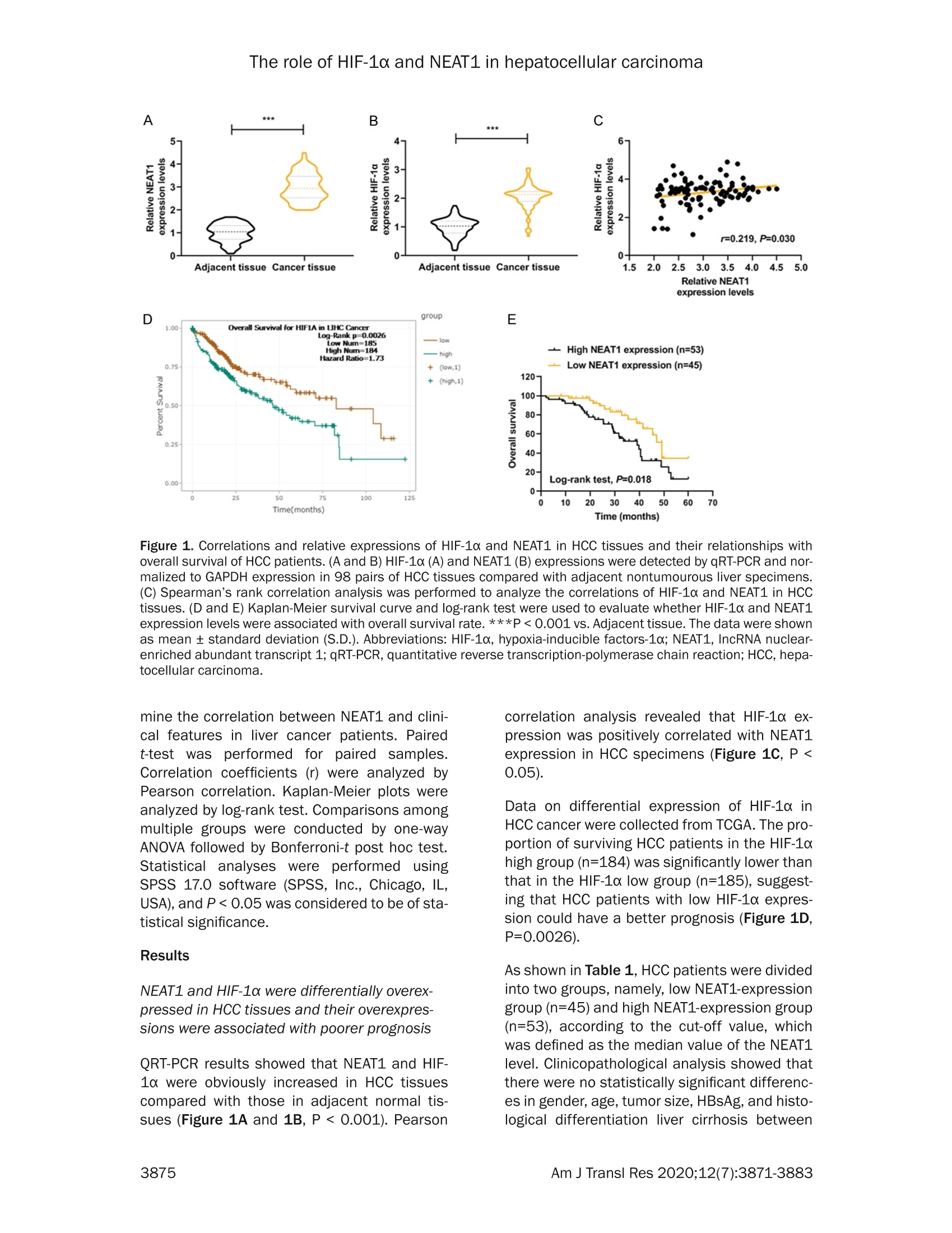
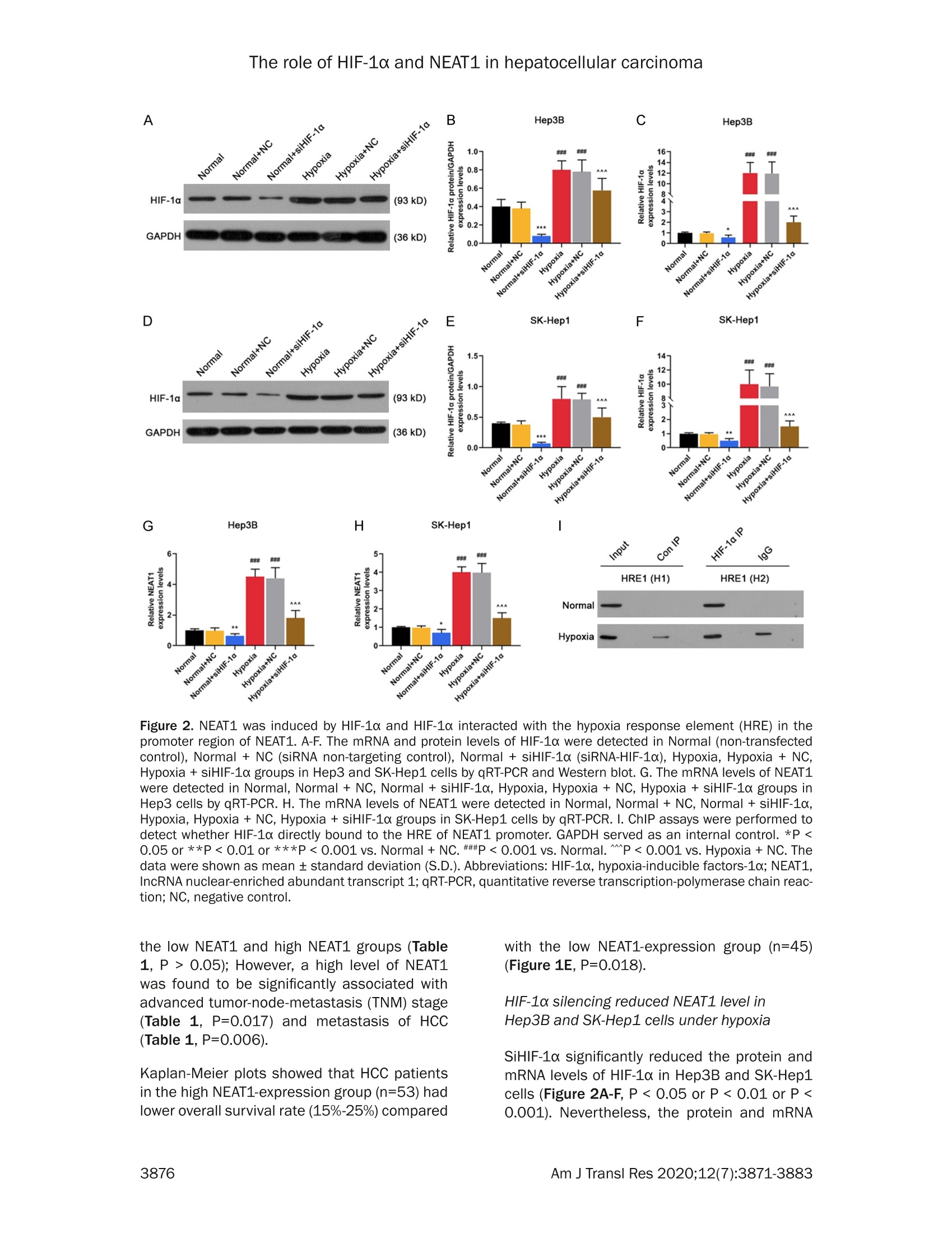

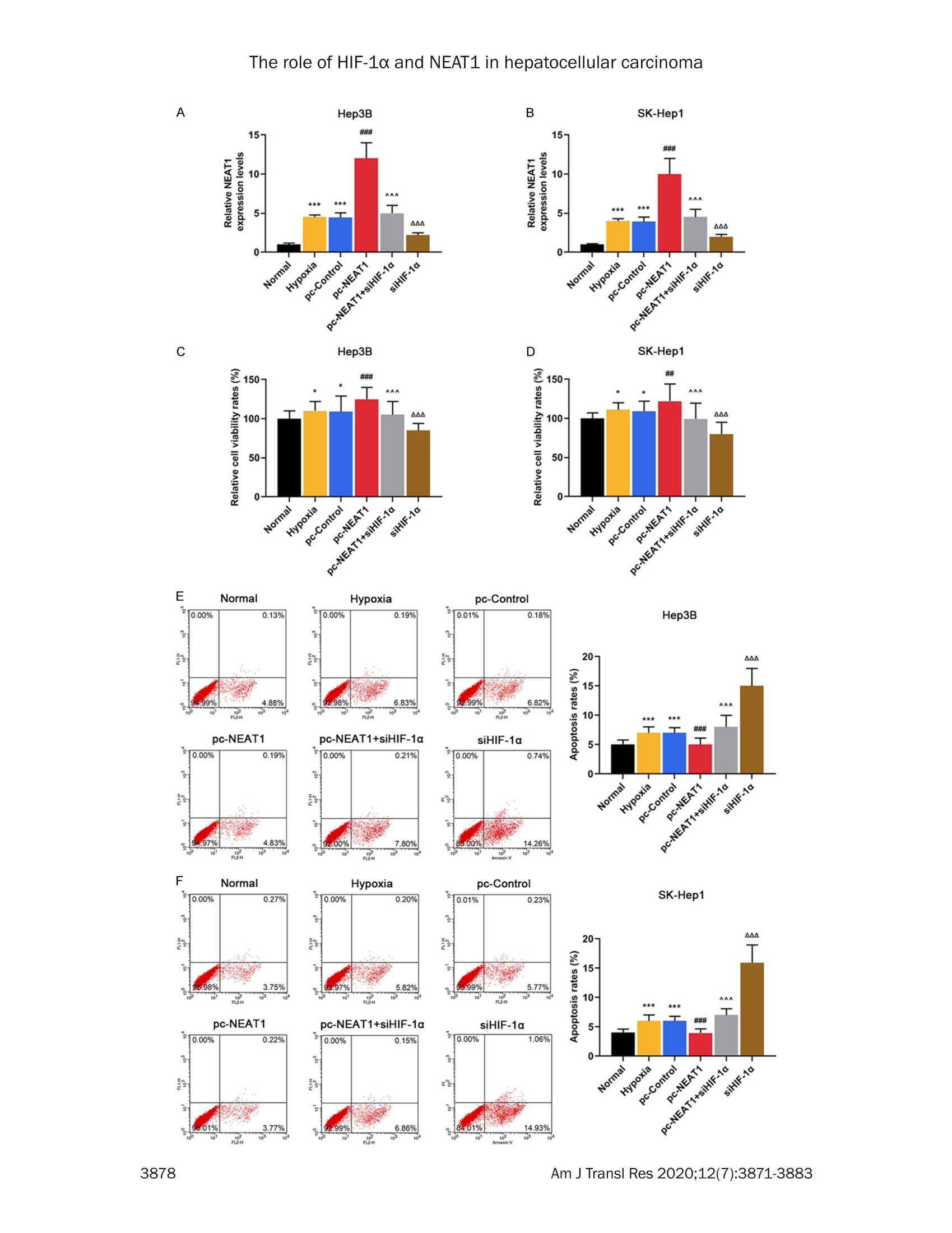



还剩11页未读,是否继续阅读?
北京隆福佳生物科技有限公司为您提供《肝细胞癌组织中缺氧诱导因子检测方案(其它培养箱)》,该方案主要用于癌细胞/肿瘤细胞中其他检测,参考标准--,《肝细胞癌组织中缺氧诱导因子检测方案(其它培养箱)》用到的仪器有
该厂商其他方案
更多









