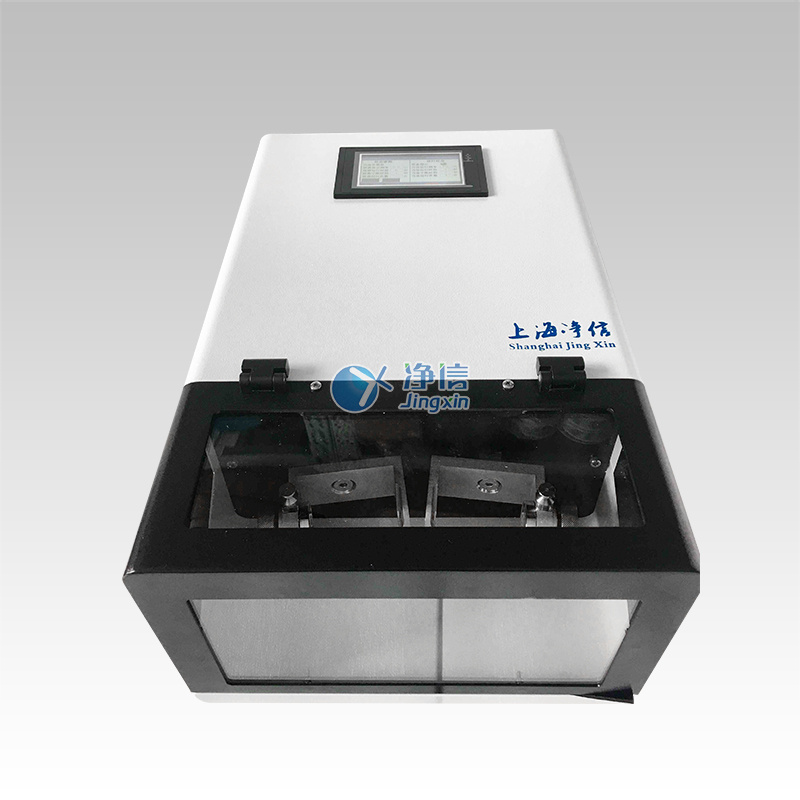
木本植物传统核酸提取方式耗时长、需使用有毒化学品(例如苯酚和氯仿),且一次只能处理少量样品。为了克服这些问题,我们开发了一种不使用苯酚/氯仿、基于CTAB / PVP的新方法,用于RNA或DNA提取。该方法同样适用于96深孔板,使用SPEX高通量组织研磨仪,一次研磨6块深孔板,同时处理576个样品,大大加快了处理速度。
方案详情

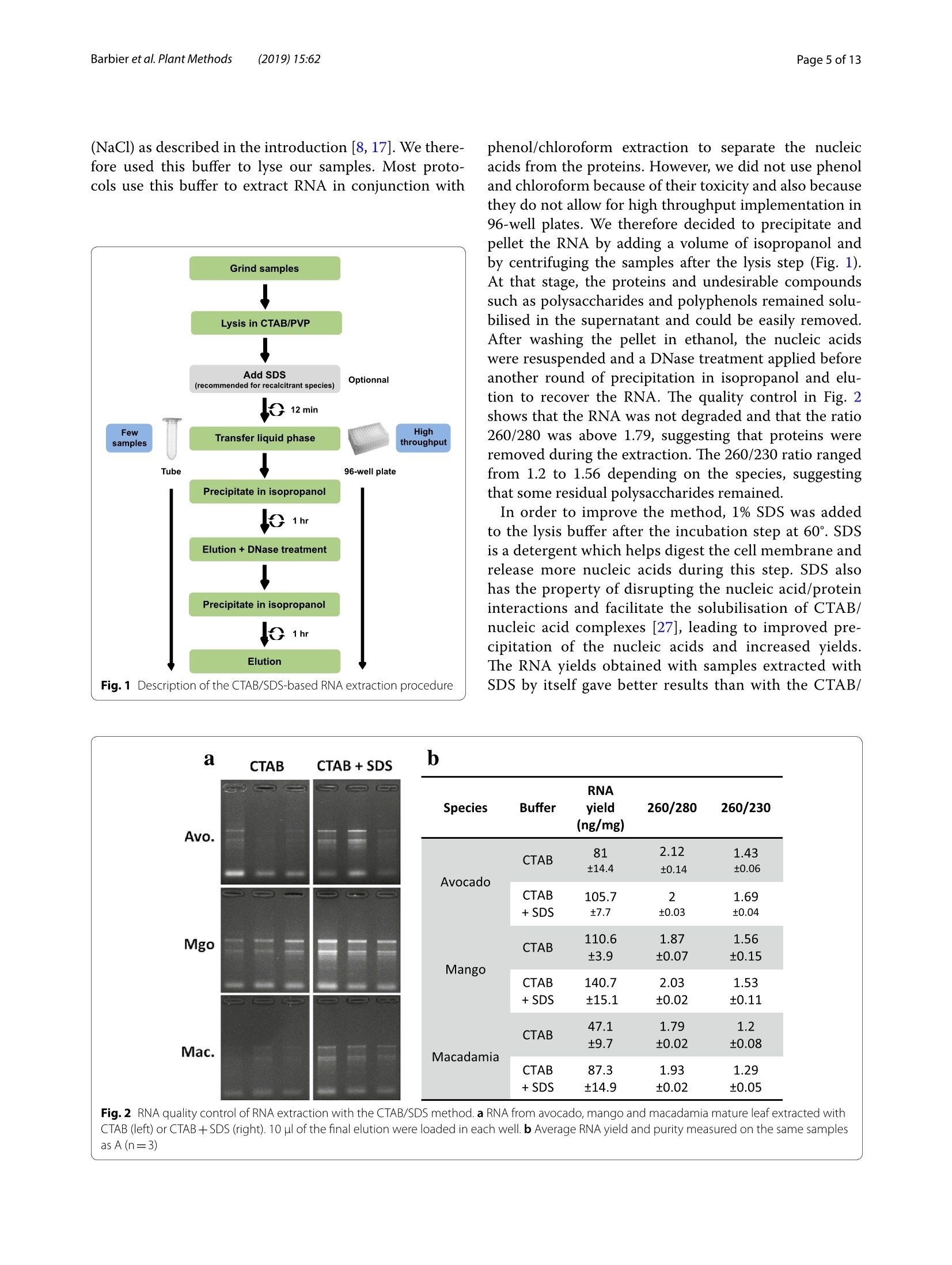
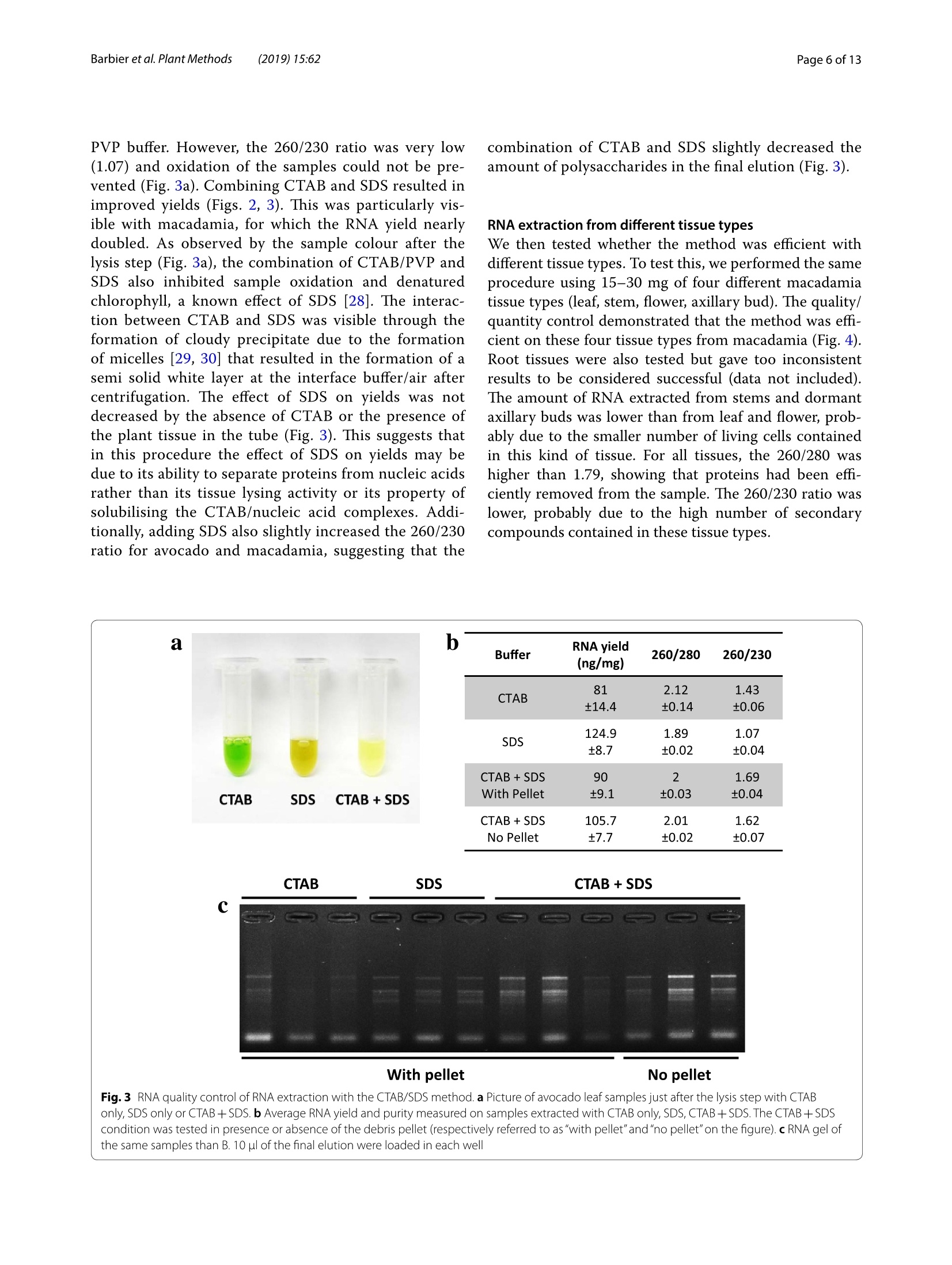
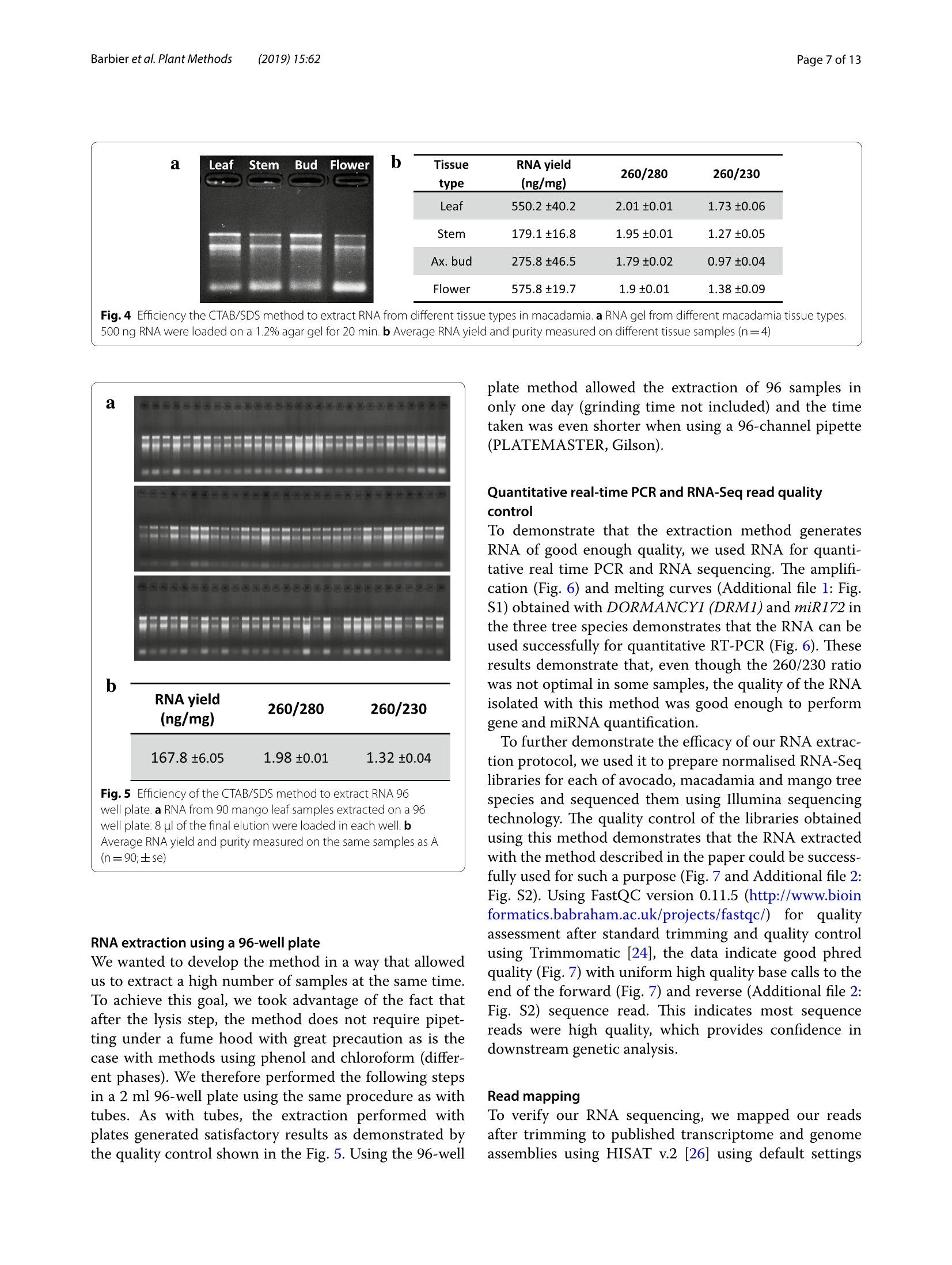
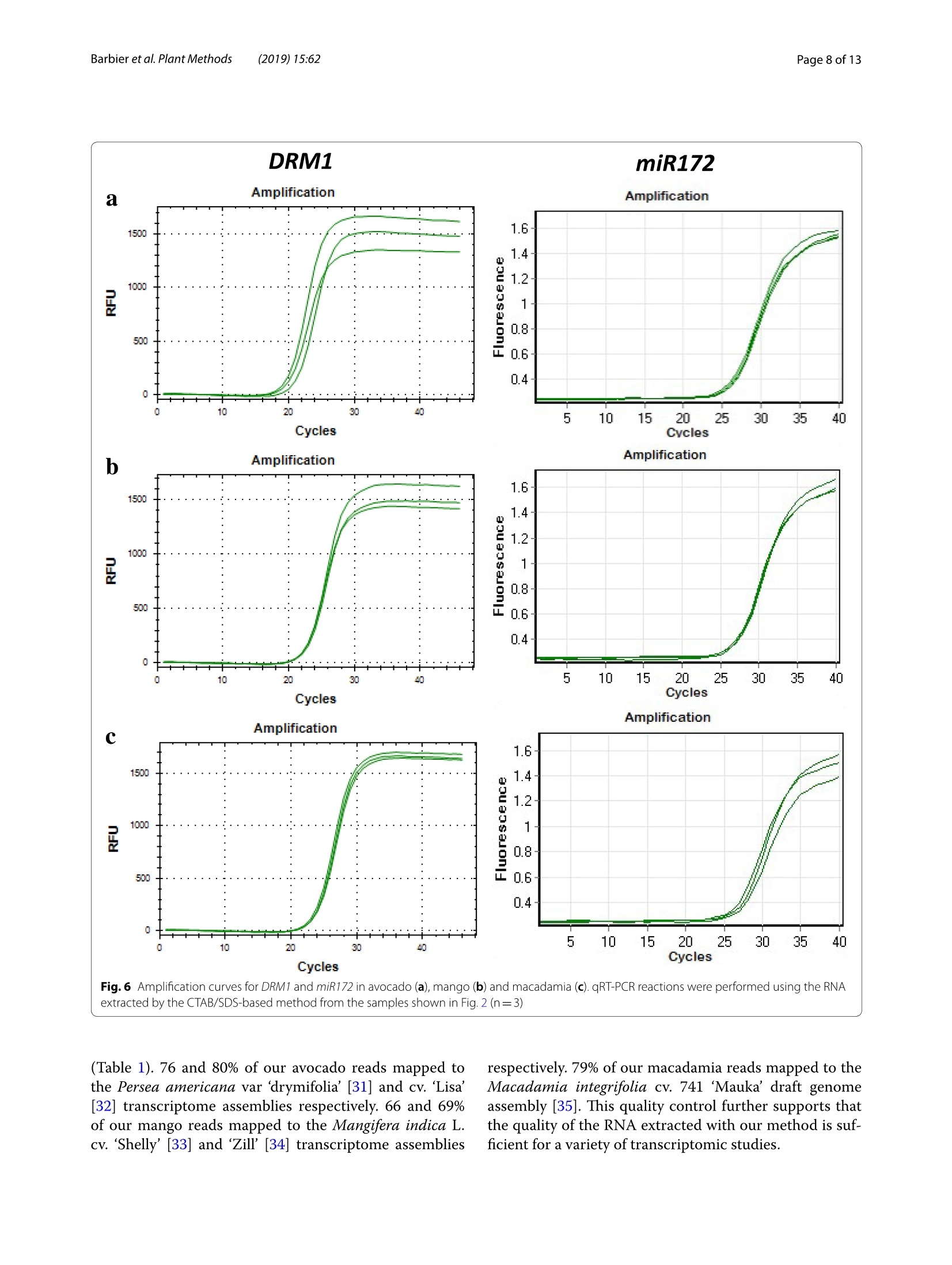
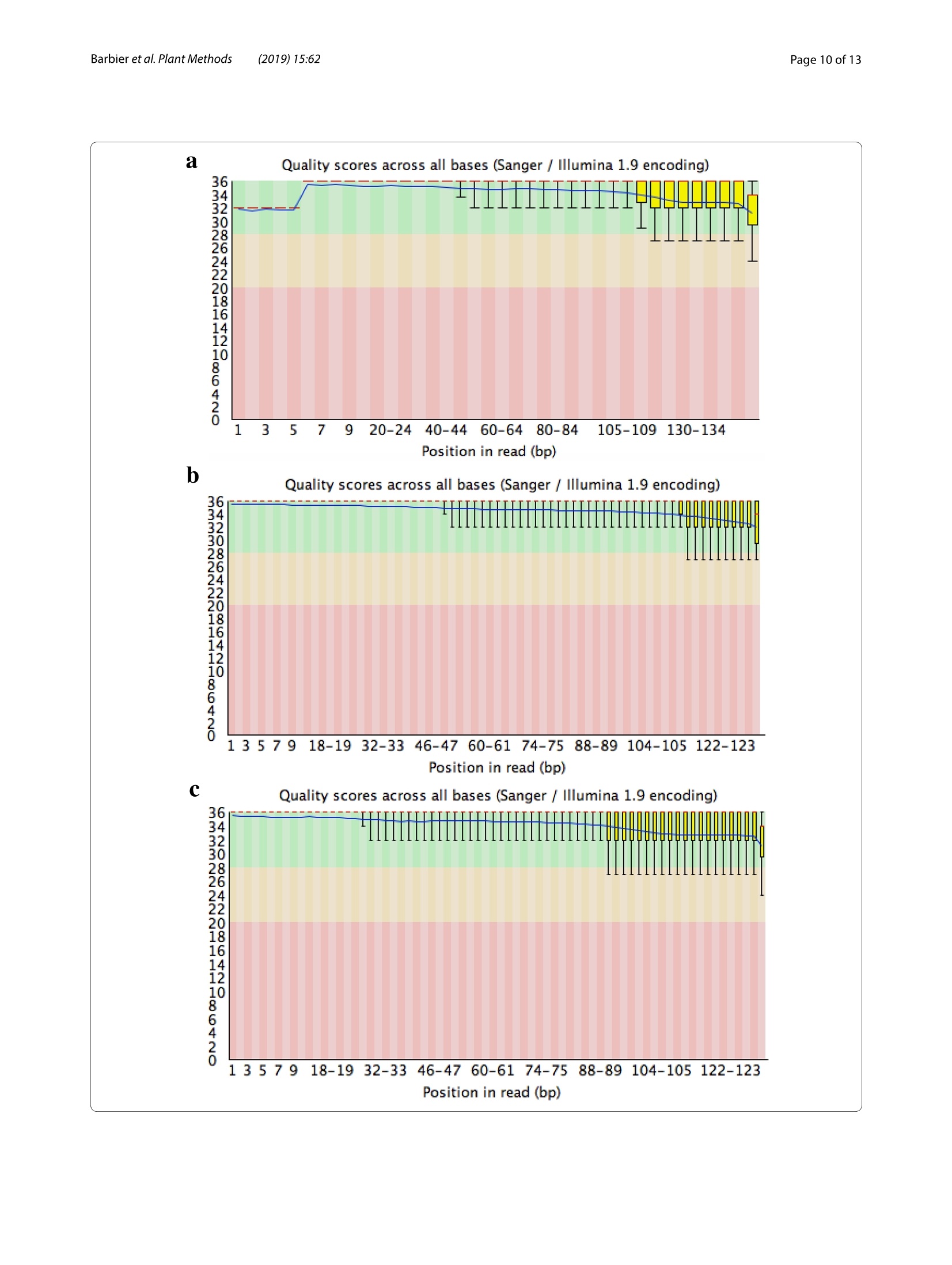
Barbieret al. Plant Methods (2019)15:62https://doi.org/10.1186/s13007-019-0447-3Plant Methods Barbier et al. Plant Methods (2019)15:62Page 2 of 13 *Correspondence: barbier.francois@hotmail.frSchool of Biological Sciences, and The Queensland Alliancefor Agriculture and Food Innovation, The University of Queensland, St.Lucia,QLD 4072,AustraliaO The Author(s) 2019. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License, (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium,provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license,and indicate if changes were made.The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated. METHODOLOGY Open Access A phenol/chloroform-free method to extractCuhpedcak tfeosrnucleic acids from recalcitrant, woody tropicalspecies for gene expression and sequencing Francois F. BarbierD,Tinashe G. ChabikwaD, Muhammad U. AhsanD, Stacey E. CookD,Rosanna PowellDMilos TanurdzicD and Christine A. BeveridgeD Abstract Background: Woody tropical plants contain high levels of complex organic compounds that inhibit the chemicalprocedures needed to extract RNA or DNA, thus compromising downstream applications such as RNA sequencingand analysis of gene expression. To overcome this issue, researchers must use extraction protocols using CTAB/PVPbuffer instead of commercially available DNA/RNA extraction kits. However, these protocols are time-consuming, usetoxic chemicals like phenol and chloroform, and can only be used to process a small number of samples at a time.To overcome these issues, we developed a new CTAB/PVP based protocol for RNA or DNA extraction that eliminatesthe traditional phenol/chloroform step. Furthermore, the protocol was developed for 96-well plates to speed upprocessing. Results: Our new protocol enabled us to successfully extract RNA from macadamia, avocado, and mango tissues thatare traditionally difficult to work with. This RNA was then successfully used to synthesise cDNA for real-time quantita-tive PCR and to generate good quality RNA-Seq libraries. Our protocol can be easily converted for rapid DNA extrac-tion from different tropical and sub-tropical tree species. Conclusion: This method enables safer and faster DNA and RNA extraction from recalcitrant species, thus facilitatingfuture work on tropical trees. Keywords: DNA RNA extraction, CTAB, SDS, Woody species, Macadamia, Avocado, Mango Background Molecular experiments reveal the physio-genetic struc-ture and function of plant species, enabling growers toimprove productivity in changing environmental con-ditions. However, the protocols used to extract RNA orDNA from plants were developed using herbaceous spe-cies such as Arabidopsis (Arabidopsis thaliana L. (Heyn))[1-4], and do not work well for some taxa. For example,the tissues of tropical trees contain polysaccharides andpolyphenols that compromise the extraction of nucleicacids [5,6]. Extraction of RNA or DNA from these species relies on the use of phenol and chloroform, whichare volatile, toxic, and therefore impractical for routineand repeated use by researchers. Therefore, we soughtto improve the extraction of RNA or DNA from tropicaltrees by creating a protocol that is safer and faster. For herbaceous plants, the extraction of RNA or DNAcan be easily achieved with silica membranes, how-ever these methods do not work efficiently with tropicalwoody species. Furthermore, this technique only retainsRNA strands longer than fifty nucleotides, eliminat-ing the small RNA which are of emerging importancein molecular plant sciences [7]. Therefore, in the cur-rent study we began with modifications to a protocolfor RNA extraction from pine tree [8], which uses cetyltrimethylammonium bromide (CTAB) and polyvinylpo-lypyrrolidone (PVP) in the lysis buffer [2, 9-12]. CTAB In this study, we developed and tested a new methodfor extracting DNA and total RNA, including small RNA,from diverse tissues of tropical/subtropical woody spe-cies, including Avocado (Persea americana L.), Macada-mia (Macadamia integrifolia L.) and Mango (Mangiferaindica L.). Our method does not require phenol or cholo-rophorm and can be used to extract 96 samples in a96-well plate in less than 1 day. The resultant RNA can besequenced and used to produce good quality RNA-Seqreads. The DNA extraction method can also be used onCoffee tree (Coffea arabica L.) and Eucalyptus (Eucalyp-tus grandis L.) samples. This protocol will facilitate novelmolecular research on tropical trees that have previ-ously been too difficult for large-scale genetic survey andexperimentation. Materials and methods Plant material and tissue grinding Various tissue samples were collected from field-grownAvocado cv. Hass, Mango cv. 1243, Macadamia cv. 751,Coffee and Eucalyptus in Queensland, Australia. All thetrees were mature (8-15 year old).DNA was extractedfrom mature leaves whilst RNA extractions were per-formed on stem, leaf, root, flower and axillary bud tis-sues. Arabidopsis cv. Columbia-0 and garden pea cv.Torsdag were also used to assess the efficiency of themethod on herbaceous plants. Fresh material was flash frozen in liquid nitrogen or dryice and stored at -80℃ before being ground to pow-der (homogenised). Frozen samples were ground withan automated tissue grinder (Geno/Grinder, SPEX).Leaf, bud, flower and root samples were ground in 2 mltubes (up to 96 at a time) and stem tissues were groundin 15 ml vials (up to 12 at a time). Frozen samples weredisrupted for 1 min at 1750 rpm and refrozen for 30 minat -80℃ before being disrupted again for one or twocycles. The amount of powder required for the extractionwas transferred in a new tube using a spatula. The esti-mated weight was assessed based on preliminary assess-ment volume/weight. Solutions and reagents 。 CTAB buffer: CTAB 2%, NaCl 1.4 M,EDTA 20 mM,Tris-HCl 100 mM, PVP40 2% Isopropanol 100% 。 DTT (DL-Dithiothreitol) 0.5 mM SDS 10% 70% ethanol 。 DNaseI+DNase buffer 。RNase A RNA extraction Lysis of Tissues · Transfer 0.5-50 mg (not more) of ground sampleinto a 2 ml tube 。 Add 625 pl of CTAB buffer and 25 pl of DTT 0.5 mM(mix them prior to use) ◆Vortex well and incubate at 60C for 15 min. Vortexevery5 min ·ONLY FOR RECALCITRANT SAMPLES: Add 65 ulof SDS 10% and vortex well (if you use small amountof tissue, only add 40 pl of SDS 10%) Note: At that stage, SDS precipitates with CTABmaking the sample cloudy 。 Centrifuge at 20,000g for 15 min at room tempera-tureNote: Tissue debris will pellet in the bottom of thetube and SDS/CTAB will form a semi-solid layer atthe surface · Remove the tubes carefully from the centrifuge andtransfer 550-600 pl of the liquid phase into a new2 ml tube or into a 2 ml cavity of a 96 well plate andproceed to the nucleic acid precipitation Note 1: In case too much top layer has been mistak-enly taken up, put the samples into new 2 ml tubesand repeat this step (tip: after this second centrifuge, take the sample by pipetting directly from the bot-tom of the tube (the top layer will stick to the out-side of the tip). Note 2: Unclear precipitate can form depending onthe tissue (this was the case with avocado buds).Transfer into a filter column (Qiagen QIAshredderor Macherey-Nagel NucleoSpin Filters) instead ofthe 2 ml tube and repeat this step. Nucleic acid precipitation · Add 550 pl of pre-cooled (-20℃) isopropanol intoeach tube/well and vortex well .Leave at -20℃ for 15 min Note: At that stage you can leave the samples fordays or weeks at -20℃ . Centrifuge the tubes/plate at 4℃ for 1 h at 3100g forplates or 45 min at 20,000g for tubes 。 Tip the tubes/plate to remove isopropanol 。Add1ml 70% ethanol .Centrifuge at 4℃ for 10-15 min at 3100g for platesor 20,000g for tubes 。 Tip the tubes/plate to remove the ethanol · Centrifuge the tubes/plate brieflyy andremoveremaining ethanol with a pipette Let the samples dry on the bench for 10 min. DNase treatment · Add 175 ul of RNase-free water and pipette up anddown to resuspend the pellet .Place the plate at 50-60℃ for 2-3 min, vortex andquick spin down Add 20 ul of DNase buffer and 5 pl of DNase I pertube or well (mix them before) Vortex briefly, quick spin down and incubate at 37℃for 20-30 min Immediately proceed to the next step. RNA precipitation and elution Add 200 ul of pre-cooled (-20℃) isopropanol intoeach tube/well and vortex well Leave for 15 min at -20℃Note: At that stage, you can leave the samples forseveral days at -20 ℃ · Centrifuge at 4 ℃ for 1 h at 3100 g for plates or45 min at 20,000g for tubes Tip the tubes/plate to remove isopropanol 。 Add1ml of 70% ethanol ·Centrifuge at 4℃ for 10-15 min at 3100 g forplates or 20,000g for tubes Tip tubes/plate to remove ethanol · Centrifuge the tubes/plate briefly and removeremaining ethanol with a pipette · Let the samples dry on the bench for 10 min · Add 40-100 ul of warm (60°℃) RNase-free waterand pipette up and down vigorously · Vortex well and quick spin down 。 Determine RNA quantity and quality by gel elec-trophoresis and spectrophotometry, and store thesamples at -80C. DNA extraction Lysis of Tissues and RNase treatment Transfer up to 50 mg (no more) of ground sampleto a 2 ml tube · Add 400 ul of CTAB buffer and 4 ul of RNaseA+RNase buffer (mix them together as before) 。Vortex well and incubate at 37 C for 1 h. Vortexthem and invert every 15 min Add 30 ul of SDS 10% and vortex well (if you use asmall amount of tissue (<10-15 mg), add only 15 plof SDS 10%) Note: SDS precipitates with CTAB making thesample cloudy · Centrifuge at 20,000g for 15 min at room tempera-tureNote: Tissue debris will pellet in the bottom ofthe tube and SDS/CTAB will form a semi-solidlayer at the surface Remove carefully from the centrifuge and trans-fer 350-400 ul of the liquid phase into a new 2 mltube or into a 2 ml 96 well plate and proceed to thenucleic acid precipitation Note 1: In case too much top layer has been mis-takenly taken up, put the samples into new 2 mltubes and repeat this step (tip: after this secondcentrifuge, take the sample by pipetting directlyfrom the bottom of the tube (the top layer willstick to the outside of the tip). Note 2: In case of dirty/unclear precipitate canform depending on the tissue (this was the casewith avocado buds), transfer into a filter column(Qiagen QIAshredder or Macherey-Nagel Nucle-oSpin Filters) instead of the 2 ml tube and repeatthis step. DNA precipitation and elution . Add 350-400 ul of pre-cooled (-20℃) isopro-panol to each tube/well and vortex well Leave at -20C for 15 min Note: At that stage, you can leave the samples forseveral days at -20°℃ · Centrifuge the tubes/plate at 4℃ for 1 h at 3100gfor plates or 45 min at 20,000g for tubes Tip the tubes/plate to remove isopropanol 。 Add1 ml of 70% ethanol Centrifuge at 4℃ for 10-15 min at 3100g for platesor 20,000g for tubes Tip the tubes/plate to remove ethanol Centrifuge the tubes/plate briefly and removeremaining ethanol with a pipette Let the samples dry on the bench for 10 min ◆· Add 90 ul of warm (60℃) RNase-free water,pipette up and down and vortex well to resuspendthe pellet. Quantification and quality control Quality control was performed by running RNA orDNA samples on a 1.1% agarose gel. The nucleic acidswere stained by adding Red Sage to the gel. Gel visu-alisation was performed using the Gel Doc Gel Docu-mentation System (Bio-Rad Laboratories, California,USA). The quality and purity of the samples were deter-mined with a NanoVuePlus Spectrophotometer (GEHealthcare Life Sciences, Pennsylvania, USA) meas-uring the absorbance at 280 nm for RNA and 260 forDNA. cDNA synthesis and quantitative real-time PCR (qRT-PCR) For qRT-PCR, cDNA was obtained by reverse tran-scription using 250-500 ng of total RNA in 8 pl and2 ul of 5xiScript Supermix (Bio-Rad Laboratories,California, USA), following the manufacturer’s instruc-tions. The cDNA was then diluted to 0.5 ng equivalentRNA in milliQ water per ul for a working templatesolution. Quantitative real-time PCR was then per-formed using 2 pl of 1 mM primer mix, 5 ul of cDNAand 3 ul SensiFASTM SYBR No-ROX Kit (Bioline) perreaction. Samples were amplified following the manu-facturer's instructions and fluorescence was monitoredwith a CFX384 Touch Real-Time PCR Detection Sys-tem (Bio-Rad Laboratories, California, USA). cDNA synthesis and miRNA expression quantification For quantification of miRNA expression, low molecu-lar weight cDNA was synthesised by ligation-mediated reverse transcription from 100 to 200 ng of total RNAusing a miScript Plant RT Kit (Qiagen, The Nether-lands) kit per manufacturer’s protocol. The preparedcDNA was then diluted with milliQ water as per themanufacturer's instructions for optimal amplification.The qRT-PCR reactions were performed as per manu-facturer’s instructions (miScript SYBR Green PCR Kit,Qiagen, The Netherlands) using: 10 ul of 1xQuanti-Tect SYBRgreen mastermix, 2 pl of 1 x miScript uni-versal primer, 2 pl0.8 uM miRNA specific primer and2 ultemplate cDNA. The qRT-PCR run was performed,and fluorescence was measured using a Rotor-Gene Q6000 (Qiagen, The Netherlands). RNA-Seq library preparation and sequencing RNA libraries of pooled avocado, macadamia and mangostem, leaf, root, flower and axillary bud tissues were pre-pared as described by Kerr et al. [23]. cDNA librarieswere then normalised using the Trimmer-2 cDNA nor-malisation kit following the manufacturer's instructions(Evrogen JSC, Moscow, Russia). Libraries were quantifiedby qRT-PCR using the Library Quantification Kit for Illu-mina sequencing platforms (KAPA Biosystems, Boston,USA), using a CFX96 Touch" Real-Time PCR Detec-tion System (Bio-Rad Laboratories, California, USA).The libraries were normalised to a working concentrationof 4 nM using the molarity calculated from qRT-PCR,adjusted for fragment size. The RNA-Seq libraires werethen sequenced using an Illumina Sequencer (NextSeq). RNA-Seq read quality control assessment and mapping Trimmomtatic v. 0.35 [24] was used to remove adaptors,trim and filter the reads based on quality. Sequence con-taminants were removed from the reads using Deconseqv. 0.4.2 [25]. Sequence read quality was checked usingFastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). To validate our RNA sequencing results, we mappedthe reads to published draft transcriptome and genomesequences. Trimmed reads were mapped to publishedreference transcriptome and genome assemblies usingHISAT v.2 [26] using default settings. Results Phenol/chloroform-freeCTAB/PVP/SDS-based RNAextraction In the first couple of experiments described below, wedeveloped and tested an RNA extraction method using40-60 mg of frozen leaf tissue from avocado, macadamiaand mango mature leaves. Most of the published RNAextraction procedures developed for recalcitrant treespecies improved the RNA yield and quality by using aCTAB/PVP lysis buffer with high concentration of salt (NaCl) as described in the introduction [8, 17]. We there-fore used this buffer to lyse our samples. Most proto-cols use this buffer to extract RNA in conjunction with Fig. 1 Description of the CTAB/SDS-based RNA extraction procedure phenol/chloroform extraction to separate the nucleicacids from the proteins. However, we did not use phenoland chloroform because of their toxicity and also becausethey do not allow for high throughput implementation in96-well plates. We therefore decided to precipitate andpellet the RNA by adding a volume of isopropanol andby centrifuging the samples after the lysis step (Fig.1).At that stage, the proteins and undesirable compoundssuch as polysaccharides and polyphenols remained solu-bilised in the supernatant and could be easily removed.11..,After washing the pellet in ethanol, the nucleic acidswere resuspended and a DNase treatment applied beforeanother round of precipitation in isopropanol and elu-tion to recover the RNA. The quality control in Fig. 2shows that the RNA was not degraded and that the ratio260/280 was above 1.79, suggesting that proteins wereremoved during the extraction. The 260/230 ratio rangedfrom 1.2 to 1.56 depending on the species, suggestingthat some residual polysaccharides remained. In order to improve the method, 1% SDS was addedto the lysis buffer after the incubation step at 60°. SDSis a detergent which helps digest the cell membrane andrelease more nucleic acids during this step. SDS alsohas the property of disrupting the nucleic acid/proteininteractions and facilitate the solubilisation of CTAB/nucleic acid complexes [27], leading to improved pre-ds:cipitation of the nucleic acids and increased yields.The RNA yields obtained with samples extracted withSDS by itself gave better results than with the CTAB/ Fig. 2 RNA quality control of RNA extraction with the CTAB/SDS method. a RNA from avocado, mango and macadamia mature leaf extracted withCTAB (left) or CTAB+SDS (right). 10 ul of the final elution were loaded in each well. b Average RNA yield and purity measured on the same samplesas A(n=3) PVP buffer. However, the 260/230 ratio was very low(1.07) and oxidation of the samples could not be pre-vented (Fig.3a). Combining CTAB and SDS resulted inimproved yields (Figs. 2, 3). This was particularly vis-ible with macadamia, for which the RNA yield nearlydoubled. As observed by the sample colour after thelysis step (Fig. 3a),the combination of CTAB/PVP andSDS also inhibited sample oxidation and denaturedchlorophyll, a known effect of SDS [28]. The interac-tion between CTAB and SDS was visible through theformation of cloudy precipitate due to the formationof micelles [29, 30] that resulted in the formation of asemi solid white layer at the interface buffer/air aftercentrifugation. The effect of SDS on yields was notdecreased by the absence of CTAB or the presence ofthe plant tissue in the tube (Fig. 3). This suggests thatin this procedure the effect of SDS on yields may bedue to its ability to separate proteins from nucleic acidsrather than its tissue lysing activity or its property ofsolubilising the CTAB/nucleic acid complexes. Addi-tionally, adding SDS also slightly increased the 260/230ratio for avocado and macadamia, suggesting that the combination of CTAB and SDS slightly decreased theamount of polysaccharides in the final elution (Fig. 3). RNA extraction from different tissue types We then tested whether the method was efficient withdifferent tissue types. To test this, we performed the sameprocedure using 15-30 mg of four different macadamiatissue types (leaf, stem, flower, axillary bud). The quality/quantity control demonstrated that the method was effi-cient on these four tissue types from macadamia (Fig.4).Root tissues were also tested but gave too inconsistentresults to be considered successful (data not included).The amount of RNA extracted from stems and dormantaxillary buds was lower than from leaf and flower, prob-ably due to the smaller number of living cells containedin this kind of tissue. For all tissues, the 260/280 washigher than 1.79, showing that proteins had been effi-ciently removed from the sample. The 260/230 ratio waslower, probably due to the high number of secondarycompounds contained in these tissue types. Fig. 3 RNA quality control of RNA extraction with the CTAB/SDS method. a Picture of avocado leaf samples just after the lysis step with CTABonly, SDS only or CTAB+SDS. b Average RNA yield and purity measured on samples extracted with CTAB only, SDS, CTAB+ SDS. The CTAB+SDScondition was tested in presence or absence of the debris pellet (respectively referred to as "with pellet"and"no pellet"on the figure). c RNA gel ofthe same samples than B. 10 ul of the final elution were loaded in each well Tissue RNA yield 260/280 260/230 type (ng/mg) Leaf 550.2±40.2 2.01±0.01 1.73±0.06 Stem 179.1±16.8 1.95±0.01 1.27±0.05 Ax. bud 275.8±46.5 1.79±0.02 0.97±0.04 Flower 575.8±19.7 1.9±0.01 1.38±0.09 Fig.4 Efficiency the CTAB/SDS method to extract RNA from different tissue types in macadamia. a RNA gel from different macadamia tissue types.500 ng RNA were loaded on a 1.2% agar gel for 20 min. b Average RNA yield and purity measured on different tissue samples (n=4) (n=90;±se) RNA extraction using a 96-well plate We wanted to develop the method in a way that allowedus to extract a high number of samples at the same time.To achieve this goal, we took advantage of the fact thatafter the lysis step, the method does not require pipet-ting under a fume hood with great precaution as is thecase with methods using phenol and chloroform (differ-ent phases). We therefore performed the following stepsin a 2 ml 96-well plate using the same procedure as withtubes. As with tubes, the extraction performed withplates generated satisfactory results as demonstrated bythe quality control shown in the Fig. 5. Using the 96-well plate method allowed the extraction of 96 samples inonly one day (grinding time not included) and the timetaken was even shorter when using a 96-channel pipette(PLATEMASTER, Gilson). Quantitative real-time PCR and RNA-Seq read qualitycontrol To demonstrate that the extraction method generatesRNA of good enough quality, we used RNA for quanti-tative real time PCR and RNA sequencing. The amplifi-cation (Fig. 6) and melting curves (Additional file 1: Fig.S1) obtained with DORMANCY1(DRM1) and miR172 inthe three tree species demonstrates that the RNA can beused successfully for quantitative RT-PCR (Fig.6). Theseresults demonstrate that, even though the 260/230 ratiowas not optimal in some samples, the quality of the RNAisolated with this method was good enough to performgene and miRNA quantification. To further demonstrate the efficacy of our RNA extrac-tion protocol, we used it to prepare normalised RNA-Seqlibraries for each of avocado, macadamia and mango treespecies and sequenced them using Illumina sequencingtechnology. The quality control of the libraries obtainedusing this method demonstrates that the RNA extractedwith the method described in the paper could be success-fully used for such a purpose (Fig. 7 and Additional file 2:Fig. S2). Using FastQC version 0.11.5 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/))for qualityassessment after standard trimming. Ianngd quality controlusing Trimmomatic [24], the data indicate good phredquality (Fig. 7) with uniform high quality base calls to theend of the forward (Fig.7) and reverse (Additional file 2:Fig. S2) sequence read. This indicates most sequencereads were high quality, which provides confidence indownstream genetic analysis. Read mapping To verify our RNA sequencing, we mapped our readsafter trimming to published transcriptome and genomeassemblies using HISAT v.2 [26] using default settings Fig.6 Amplification curves for DRM1 and miR172 in avocado (a), mango (b) and macadamia (c). qRT-PCR reactions were performed using the RNAextracted by the CTAB/SDS-based method from the samples shown in Fig. 2 (n=3) (Table 1). 76 and 80% of our avocado reads mapped tothe Persea americana var drymifolia'[31] and cv. ‘Lisa'[32] transcriptome assemblies respectively. 66 and 69%of our mango reads mapped to the Mangifera indica L.cv. ‘Shelly’[33] and ‘Zill [34] transcriptome assemblies respectively. 79% of our macadamia reads mapped to theMacadamia integrifolia cv. 741 ‘Mauka’ draft genomeassembly [35]. This quality control further supports thatthe quality of the RNA extracted with our method is suf-ficient for a variety of transcriptomic studies. (See figure on next page.) Fig.7 Sequence quality of RNA extracted using the CTAB/SDS RNA extraction protocol. RNA was assessed by producing high-throughput RNAsequencing (RNA-seq) libraries of the avocado, macadamia and mango samples shown in Figs. 1, 2, 3,4 and 5. The RNA-Seq libraries were pair-endsequenced (150 bp) on the Illumina 2500 Hi-Seq Platform. Sequencing quality assessment using FastQC version 0.10.1 [9] is represented in graphsdescribing quality across all bases from every sequence read at each position (a Avocado; b Mango; c Macadamia, respectively). Sequence qualityis based on phred scores, an exponential scale where, for example, 20=one incorrect sequence base-call in 100, and 30= one incorrect base-call in1000. The y-axis shows the quality scores, and the higher the score, the greater confidence in the base-calls at that position. The background of thegraph divides the y-axis into very good quality calls (green), reasonable quality (orange), and poor quality (red). The graphs are representative of theforward reads (for reverse reads, see Additional file 2: Fig.S2) Phenol/chloroform-free CTAB/PVP/SDS-based DNAextraction The issues usually met with when performing DNAextraction methods for tree species are the same as withRNA extraction methods. We therefore decided to testwhether this RNA extraction method could be convertedto a genomic DNA extraction method. To achieve this,we modified the method by replacing the DTT from theCTAB buffer with RNase in order to remove the RNAduring the lysis step. As for RNA extraction, 1% SDS wasadded at the end of the lysis step and a single precipita-tion/washing/elution round was performed to recoverthe purified DNA (Fig.8). We also included mature leafsamples from outdoor grown coffee tree and eucalyptus,two species known to be challenging in terms of DNAextraction [9]. The quality and quantity control resultspresented in Fig. 9 show that the yields obtained are sat-isfactory and that the DNA extracted was not degraded,thus demonstrating that this method could be success-fully converted into a DNA extraction method for recal-citrant tropical and subtropical species. Discussion A safer version of the CTAB/PVP-based extraction methodsSuccessful nucleic acid extraction from species recal-citrant in molecular biology requires the use of CTAB/PVP-based buffers. The results we described here cleardemonstrated that this buffer was efficient to extractRNA and DNA from different tropical and subtropicalspecies using challenging for this type of experiment. Wealso highlight that SDS-based buffer, used in some pro-cedures [36, 37], cannot subsidise the CTAB/PVP bufferwithout leading to an undesirable browning of the tissue(oxidation) companied with low 260/230 ratio. Although the CTAB/PVP-based methodss are giv-ing good results in term of RNA yield and quality, thesemethods not compatible with the increasing concern forhealth safety in the scientific community, due to the largeamounts of phenol and chloroform used in these meth-ods.Our results show that it is possible to precipitateRNA directly after the lysis step. However, RNA yields are low, especially for macadamia, a crop underrepre-sented in molecular studies. Although, CTAB and SDSreact together to form micelles, adding 1% SDS at the endof the lysing step with a CTAB/PVP-based buffer couldincrease the RNA yields. This increase in final RNA yieldswas likely to be due to the property of SDS to separateproteins from RNA, since the effect of SDS on yield didnot rely on the presence of CTAB or plant tissue in thereaction (Fig. 3). High recovery yields with small amounts of tissue In this experiment described in the Fig.4, the RNA yieldsfor leaves from recalcitrant species were higher than inthe experiment described in Fig. 2 and were quite com-parable to the yields extracted from Arabidopsis or pealeaves (Additional file 3: Fig. S3). Additionally, the ratio260/230 was also improved compared to the experimentin Fig. 2. These differences may be explained by the dif-ferent amounts of fresh tissues which were used in thesetwo experiments and suggest that this method is moreefficient when performed with less fresh tissue. Interest-ingly, the results obtained on single pea buds (less than0.5 mg) demonstrate that RNA recovery is very high(Additional file 3: Fig. S3). This suggests that this pro-cedure gives better results with small amounts of tissue(less than 25 mg) and that this method can be used torecover high RNA yield, which is an advantage for sam-ples for which on small amount of tissue can be collected. Facilitating molecular studies in orphan crops Avocado, macadamia and mango can be classified asorphan crops as they are agriculturally important yetthey are not well studied and therefore have limitedgenetic and genomic resources [38]. The impractical-ity and unsafety of the published nucleic acid extractionmethods working with these crops is likely to contributeto this effect. The methods we developed in this studydemonstrate that this simple and safer procedure allowsto extract quality RNA for qRT-PCR from mRNA andalso microRNA which are of emerging importance in a Quality scores across all bases (Sanger / Illumina 1.9 encoding) 36 13579118-1932-3346-47 60-6174-7588-89104-105122-123 Position in read (bp) Quality scores across all bases (Sanger / Illumina 1.9 encoding) 00000000 18-19 32-33 46-4760-6174-7588-89 104-105122-123 1357918-19 32-33 46-4760-6174-7588-89 104-105122-123 Position in read (bp) molecular plant biology [7]. Moreover, the methoddescribed here is compatible with sequencing technol-ogy. This aspect is important since transcriptomic anal-ysis are now routinely performed in laboratories. Ourmethod will hopefully contribute to bridging the gapbetween model crops and these orphan crops. Conclusion Within the last decade, the price of transcriptomesequencinghas decreased considerably, promptingresearchers to design larger scaled experiments. How-ever, the toxicity and impracticality of the chemicals usedfor nucleic acids extraction for woody tropical speciesrepresent a major issue for these large-scale experiments.The method we described here constitutes a solution toalleviate these issues and will prompt for more large-scale experiments led on species considered recalcitrantin molecular biology. Table 1 List of RNA-seq samples and the percentage and number of mapped reads to different reference transcriptomes Species Number of reads Number % of reads NCBI Project No. Contigs References of mapped mapped intranscriptome reads orgenome Avocado 160,082,107 122,046,598 76.24 PRJNA282441 83,650 Ibarra-Laclette et al.[31] Avocado 160,082,107 128,289,801 80.14 PRJNA391003 151,545 Liu et al. [32] Mango 197,535,231 122,456,877 65.85 PRJNA227243 57,544 Luria et al. [33] Mango 197,535,231 136,367,200 69.03 PRJNA234455 54,207 Wu et al.[34] Macadamia 159,049,565 126,873,838 79.77 PRJEB13765 23,452 Nock et al.[35] Species DNA yield 260/280 260/230 (ng/mg) Avo. 720±189 1.7±0.07 0.96±0.04 Mac. 977±126 1.56±0.03 0.76±0.04 Mgo 669±41 1.64±0.07 0.93±0.08 Coff. 678 ±179 1.63±0.05 0.71±0.02 Eucal. 703±174 1.6±0.06 0.88±0.04 Genomic DNA was extracted from 20 to 35 mg of mature leaf tissuefrom avocado, macadamia, mango, coffee and eucalyptus trees usinga modified version of the procedure. a The quality was assessed byelectrophoresis (500 ng DNA loaded on a gel). b Quantity and puritycontrol obtained on tree replicates±standard error Additional files Additional file 1: Fig. S1. Melting curves for DRM1 and miR172 inavocado (A),mango (B) and macadamia (C). qRT-PCR reactions wereperformed using the RNA extracted by the CTAB/SDS-based method fromthe samples shown in Figs.2and6(n=3). Additional file 2: Fig. S2. Sequence quality of RNA extracted using the CTAB/SDS RNA extraction protocol. RNA was assessed by producinghigh-throughput RNA sequencing (RNA-seq) libraries of the avocado,macadamia and mango samples shown in Figs. 1, 2, 3, 4 and 5. The RNA-Seq libraries were pair-end sequenced (150 bp) on the Illumina 2500 Hi-Seq Platform. Sequencing quality assessment using FastQC version 0.10.1[9] is represented in graphs describing quality across all bases from everysequence read at each position (A-Avocado; B-Mango; C-Macadamia,respectively). Sequence quality is based on phred scores, an exponential scale where, for example, 20=one incorrect sequence base-call in 100,and 30=one incorrect base-call in 1000. The y-axis shows the qualityscores, and the higher the score, the greater confidence in the base-callsat that position. The background of the graph divides the y-axis into verygood quality calls (green), reasonable quality (orange), and poor quality(red). The graphs are representative of the reverse reads (for forward reads, see Fig.7). Additional file 3: Fig.S3. Efficiency the CTAB/SDS method to extract RNAfrom different tissue types in Arabidopsis and pea. Average RNA yield andpurity measured on different tissue samples (data are average ±standarderror; n=3-4, or 8 for single buds). Single pea buds are individual dor-mant axillary buds (less than 0.5 mg each). Abbreviations CTAB:cetyltrimethylammonium bromide; PVP: polyvinylpyrrolidone;SDS:sodium dodecyl sulfate; DTT: dithiothreitol; EDTA: ethylenediaminetetraaceticacid. Acknowledgements We would like to thank Annette Dexter for valuable discussions about themethod and Amanda Niehaus for her help with the manuscript. We wouldalso like to thank Helen Hoffman, John Wilkie, lan Bally, Siegrid Parfitt, JarradGriffin, Hanna Toegel, Natalie Dillon, Paula Ibell and Anahita Mizani for collect-ing and providing the samples for this work. Authors'contributions FFB developed the method, contributed to the design of the project andwrote the manuscript. TGC created the cDNA assemblies, made the Fig. 7 andTable 1 and participated to the manuscript writing. MUA made the Fig.6 andparticipated to the manuscript writing. SEC made the Fig.5 and edited themanuscript. RP assisted FFB and edited the manuscript. MT contributed to thedesign of the project, data analysis, and edited the manuscript. CAB super-vised and contributed to the design of the project and edited the manuscript.All authors read and approved the final manuscript. Funding This work was financially supported by the Australian Research Council, theQueensland Government and the Horticulture Innovation Australia Limited. Availability of data and materials Not applicable. Ethics approval and consent to participate Not applicable. Consent for publication Not applicable. Competing interests The authors declare that they have no competing interests. Received: 26 April 2019 Accepted: 27 May 2019 Published online: 05 June 2019 References 1. Edwards K, Johnstone C, Thompson C. A simple and rapid method for thepreparation of plant genomic DNA for PCR analysis. Nucleic Acids Res.1991;19:1349. .2. Kasajima 1. Protocol for DNA extraction from any plant species (alkalinePVPP method).2017 [cited 2018 Jun 13]; Available from: https://www.nature.com/protocolexchange/protocols/5359#/procedure. .3 Berendzen K, Searle l, Ravenscroft D, Koncz C, Batschauer A, Coupland G,et al. A rapid and versatile combined DNA/RNA extraction protocol andits application to the analysis of a novel DNA marker set polymorphicbetween Arabidopsis thaliana ecotypes Col-0 and Landsberg erecta. PlantMethods.2005;1:4. 4 Zou Y, Mason MG, Wang Y, Wee E, Turni C, Blackall PJ, et al. Nucleic acidpurification from plants, animals and microbes in under 30 seconds. PLoSBiol.2017;15:e2003916. 5 , Schneiderbauer A, Sandermann H, Ernst D. Isolation of functionalRNA from plant tissues rich in phenolic compounds. Anal Biochem.1991;197:91-5. 6 Katterman FR,Shattuck VI. An Effective method of DNA isolation fromthe mature leaves of Gossypium species that contain large amounts ofphenolic terpenoids and tannins. Prep Biochem. 1983;13:347-59. / DArio M, Griffiths-Jones S, Kim M. Small RNAs: big impact on plant devel-opment. Trends Plant Sci. 2017;22:1056-68. 8 Chang S, Puryear J, Cairney J. A simple and efficient method for isolatingRNA from pine trees. Plant Mol Biol Rep.1993;11:113-6. .9 Healey A, Furtado A, CooperT, Henry RJ. Protocol: a simple method forextracting next-generation sequencing quality genomic DNA fromrecalcitrant plant species. Plant Methods.2014;10:21. 10.SuzukiY, Mae T, Makino A. RNA extraction from various recalcitrant planttissues with a cethyltrimethylammonium bromide-containing buffer fol-lowed by an acid guanidium thiocyanate-phenol-chloroform treatment.Biosci Biotechnol Biochem. 2008;72:1951-3. 11.Ma Z, Huang B,Xu S, Chen Y, Li S, Lin S. Isolation of high-quality total RNAfrom Chinese fir (Cunninghamia lanceolata (Lamb.) Hook). PLoS ONE.2015;10:e0130234. 12.Xu J, Aileni M, Abbagani S, Zhang P. A reliable and efficient method fortotal rna isolation from various members of spurge family (Euphorbi-aceae). Phytochem Anal.2010;21:395-8. 13.Darby GK, Jones AS, Kennedy JF, Walker RT. Isolation and analysis of thenucleic acids and polysaccharides from Clostridium welchii. J Bacteriol.1970;103:7. 14.Tummler B, Stanke F. Genomic DNA: purification. John Wiley & Sons Ltd,editor. eLS [Internet].Chichester: Wiley; 2018 [cited 2018 Jun 13]. p.1-5.Available from: http://doi.wiley.com/10.1002/9780470015902.a0005330.pub3. 15.Porebski S, Bailey LG, Baum BR. Modification of a CTAB DNA extraction. protocol for plants containing high polysaccharide and polyphenolcomponents. Plant Mol Biol Rep. 1997;15:8-15. 16.. John ME. An efficient method for isolation of RNA and DNA from plantscontaining polyphenolics. Nucleic Acids Res. 1992;20:2381. 17.Jones AS. The isolation of bacterial nucleic acids using cetyltrimethylam-. monium bromide (Cetavlon). Biochim Biophys Acta. 1953;10:607-12. 18.FFang G, Hammar S, Grumet R. A quick and inexpensive method forremoving polysaccharides from plant genomic DNA. Biotechniques1992;13(52-4):56. 19.Mason MG, Schmidt S. Research note: rapid isolation of total RNAandgenomic DNA from Hakea actities. Funct Plant Biol. 2002;29:1015. 20.Sahu SK, Thangaraj M, Kathiresan K. DNA extraction protocol for plantswith high levels of secondary metabolites and polysaccharides withoutusing liquid nitrogen and phenol. ISRN Mol Biol [Internet]. 2012 [cited2019 Apr 21];2012. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4890884/. 21. NTP (National Toxicology Program). Report on Carcinogens,FourteenthEdition [Internet]. U.S. Department of Health and Human ServicesResearch Triangle Park; 2016. Available from:http://ntp.niehs.nih.gov/go/roc14. 22.Rodrigues P, Venancio A, Lima N. Toxic reagents and expensive equip-ment: are they really necessary for the extraction of good quality fungalDNA? Lett Appl Microbiol. 2018;66:32-7. 23.Kerr SC, Gaiti F, Beveridge CA, Tanurdzic M. De novo transcriptome assem-bly reveals high transcriptional complexity in Pisum sativum axillary budsand shows rapid changes in expression of diurnally regulated genes. BMCGenom. 2017;18:221. 24.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illu-mina sequence data. Bioinformatics. 2014;30:2114-20. 25. Schmieder R, Edwards R. Fast identification and removal of sequencecontamination from genomic and metagenomic datasets. PLoS ONE.2011;6:e17288. 26.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with lowmemory requirements. Nat Methods.2015;12:357-60. ( 27 . G ani S A, Chattoraj DK, Mukhe r jee DC. Solubilization and binding of DNA- C TAB c o mplex with SD S in aq u eous med i a. Indian J B i ochem Biophys. 1999;36:233-9. ) ( 28.Smith EL. Th e action of sodium d o decyl su l fate on the chlorophyll-pro-tein compound of the spinach leaf. J G e n Ph y siol. 1941;24:583-96. ) ( 29 T ah B, Pal P, Mahato M, Talapatra GB. Aggregation behavior of SDS/C T ABcatanionic surfactant mixture in aqueous sol u tion and at the air/water interface. J Phys Chem B. 2011;115:8493-9. ) 30.Alam MS, Ragupathy R, Mandal AB. The self-association and mixedmicellization of an anionic surfactant, sodium dodecyl sulfate, and acationic surfactant, cetyltrimethylammonium bromide: conductometric,dye solubilization, and surface tension studies. J Dispers Sci Technol.2016;37:1645-54. 31.I1barra-Laclette E, Mendez-Bravo A, Perez-Torres CA, Albert VA, Mockaitis K,Kilaru A, et al. Deep sequencing of the Mexican avocado transcriptome,an ancient angiosperm with a high content of fatty acids. BMC Genom.2015;16:599. 32.Liu L, Shu B, Jue D, Wang Y, Wei Y, Shi S. Avocado fruit pulp transcrip-tomes in the after-ripening process. Not Bot Horti Agrobot Cluj-Napoca.2018;47:308-19 33.Luria N, Sela N, Yaari M, Feygenberg O, Kobiler l, Lers A, et al. De-novoassembly of mango fruit peel transcriptome reveals mechanisms ofmango response to hot water treatment. BMC Genom. 2014;15:957. 34 Wu H, Jia H, Ma X, Wang S, Yao Q,Xu W, et al. Transcriptome and pro-teomic analysis of mango (Mangifera indica Linn) fruits. J Proteomics.2014;105:19-30. ( 35 . Nock CJ, B aten A, Barkla BJ, Furtado A , Henry RJ, King GJ. Genome and t ranscriptome sequencing characterises the gene space of Macadamia i ntegrifolia (Proteaceae). BM C Genom.2016;1 7 :937. ) 36.Huded AKC, Jingade P, Mishra MK. A rapid and efficient SDS-based RNAisolation protocol from different tissues of coffee. 3 Biotech [Internet].2018 [cited 2019 Apr 25];8. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5851945/. 37.Csaikl UM, Bastian H, Brettschneider R,Gauch S, Meir A, Schauerte M, et alComparative analysis of different DNA extraction protocols: a fast, univer-. sal maxi-preparation of high quality plant DNA for genetic evaluation andphylogenetic studies. Plant Mol Biol Rep. 1998;16:69-86. 38.Varshney RK, Ribaut J-M, Buckler ES,Tuberosa R, Rafalski JA, LangridgeP. Can genomics boost productivity of orphan crops? Nat Biotechnol.2012;30:1172-6. Publisher's Note Springer Nature remains neutral with regard to jurisdictional claims in pub-lished maps and institutional affiliations. ( Ready to submit your research? Choose BMC and benefit from: ) · fast, convenient online submission · thorough peer review by experienced researchers in your field · rapid publication on acceptance ( · s u pp ort for research d a ta, i n cluding lar ge and comp l e x da t a ty p es ) ( · g old Ope n Access which fo s ters wider col l aborati o n and increased citations ) ( · m a ximum visibilit y for your researc h :over 1 0 0M website views per year ) At BMC, research is always in progress. Learn more biomedcentral.com/submissions BMC 木本热带植物中含有大量复杂的有机化合物,这些有机化合物会抑制提取RNA或DNA所需的化学程序,从而不利于后续研究及应用,如RNA测序和基因表达分析。为了克服这个问题,研究人员必须使用CTAB / PVP缓冲液的提取方案,而不能使用市售的DNA / RNA提取试剂盒。但是,这种提取方式耗时长、需使用有毒化学品(例如苯酚和氯仿),并且一次只能处理少量样品。为了克服这些问题,我们开发了一种基于CTAB / PVP的新方法,用于RNA或DNA提取,去掉了传统方法中的苯酚/氯仿步骤。此外,该方法同样适用于96孔深孔板,使用SPEX高通量组织研磨仪,一次研磨6块深孔板,同时处理576个样品,大大加快了处理速度。 我们的新方法,能够从夏威夷果、牛油果和芒果组织中,成功提取RNA,这些组织使用传统方法很难提取到RNA。而后我们还验证了,该方法提取的RNA成功地合成cDNA,以进行实时定量PCR并生成高质量的RNA-Seq库。该方法经轻微改动后,可用于从不同的热带和亚热带树种中快速提取DNA。 这种方法可以更安全、更快速地从困难样品中提取DNA和RNA,从而为将来在热带树木上的研究提供了便利。
确定








还剩11页未读,是否继续阅读?
产品配置单

培安有限公司为您提供《牛油果树、夏威夷果树、芒果树的茎、叶、根、花等样品中RNA、DNA检测方案(研磨机)》,该方案主要用于生物农业中RNA、DNA检测,参考标准--,《牛油果树、夏威夷果树、芒果树的茎、叶、根、花等样品中RNA、DNA检测方案(研磨机)》用到的仪器有HG-600 Geno/Grinder (原SPEX 2010)高通量动植物组织研磨机
推荐专场
相关方案
更多
该厂商其他方案
更多