方案详情
文
HIF(缺氧诱导因子)基因家族成员在氧利用率变化期间作为细胞和全身氧稳态的主要调节者。青藏高原是形成长期缺氧和冷适应天然实验室。在这种情况下,限于3500 m 高海拔淡水河流的硬鳍石斑鱼被选为模型,以与平原的代表性物种大鳞石斑鱼进行比较。我们克隆了不同的HIF-α,并进行从无脊椎动物到脊椎动物的系统发育分析,以鉴定HIF-α基因并分析其进化史。有趣的是,HIF-α经历了基因复制,这可能是由于进化过程中的全基因组复制(WGD)事件。PAML(发育分析软件)分析表明HIF-1αA 在三鳃鱼谱系中受到正向选择作用;为了研究高原和平原鱼类低氧适应与肿瘤抑制蛋白pVHL 调节HIF-α稳定性的关系,进行了一系列实验。比较HIF-α的荧光素酶转录活性和蛋白水平以及HIF-αs 与pVHL 的不同相互作用,显示了高原鱼和平原鱼之间的明显差异。通过pVHL 蛋白突变进行的功能验证表明,这些突变增加了HIF-α的稳定性及其对芳香烃受体核转位器ARNT 的异源二聚化亲和力。我们的研究表明,pVHL 的错义突变诱导了生活在高海拔低氧环境中的高原鳅的进化分子适应。
方案详情

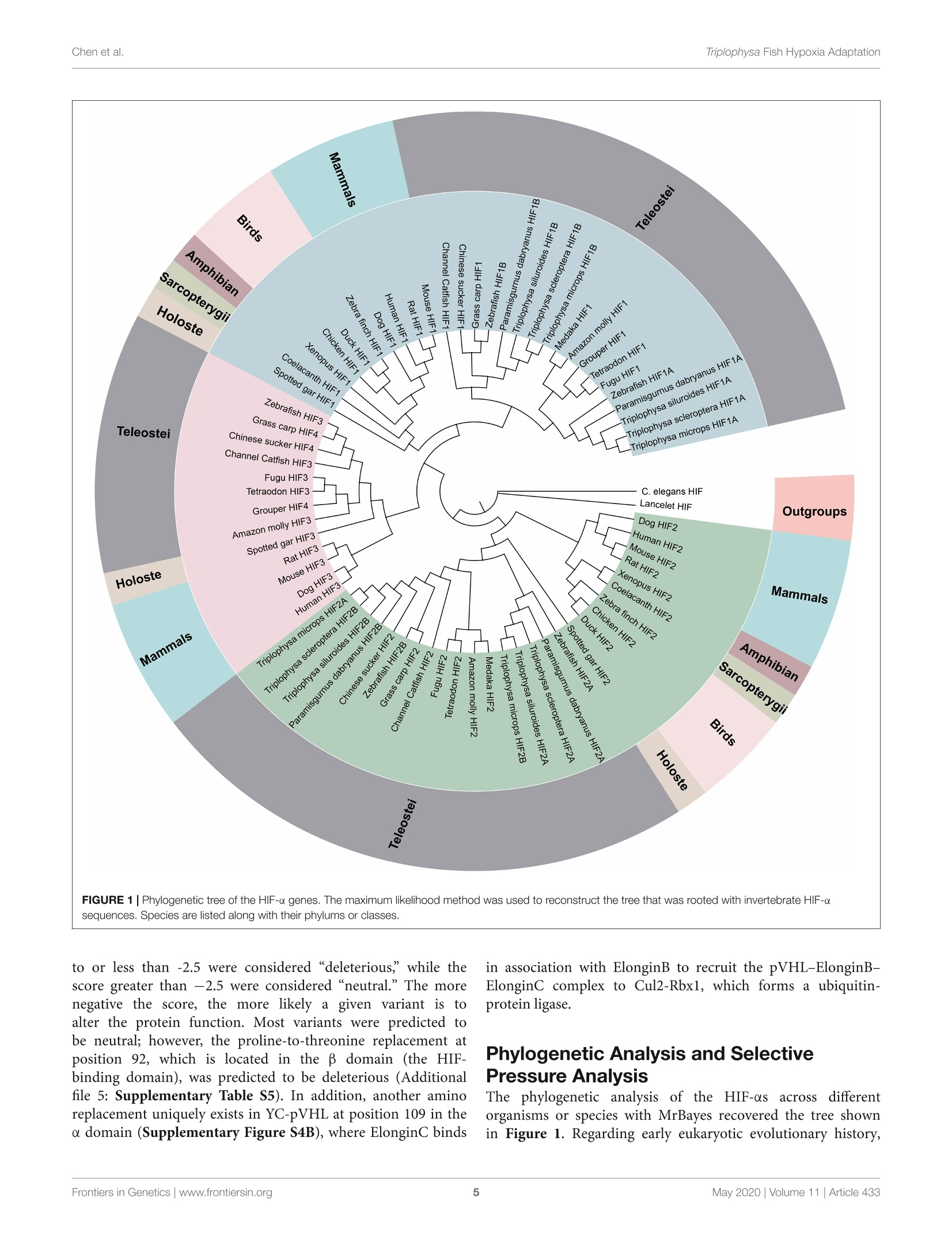
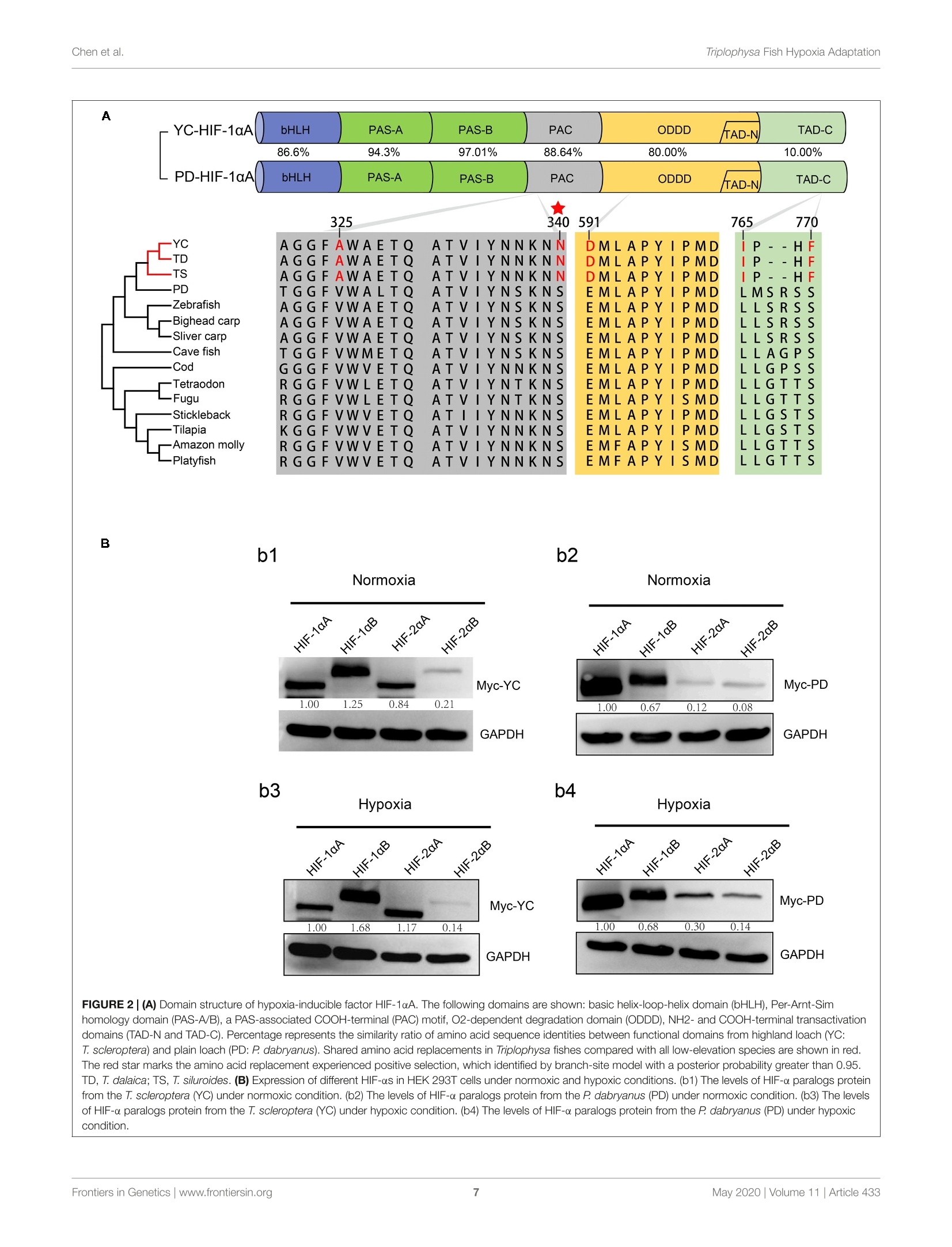
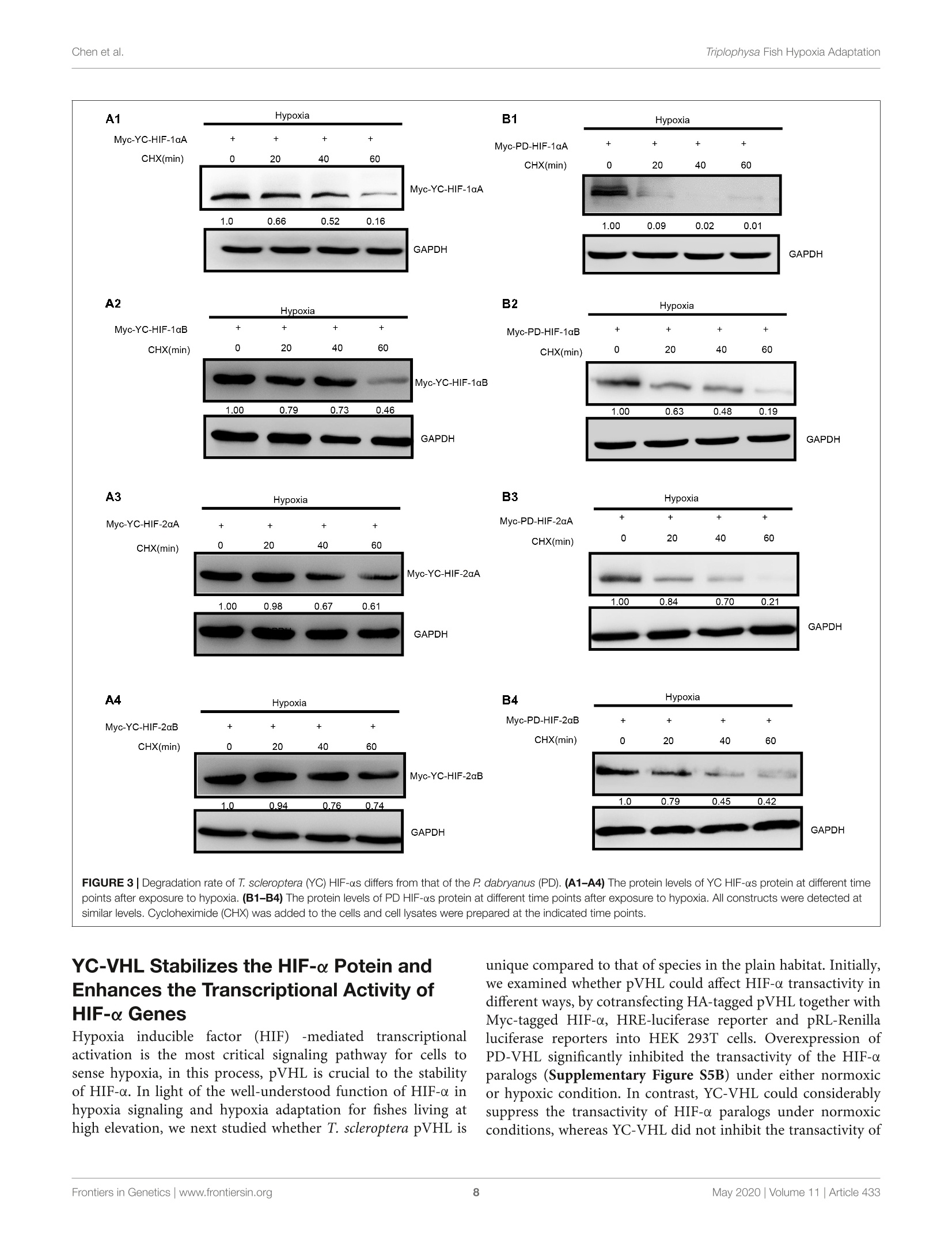
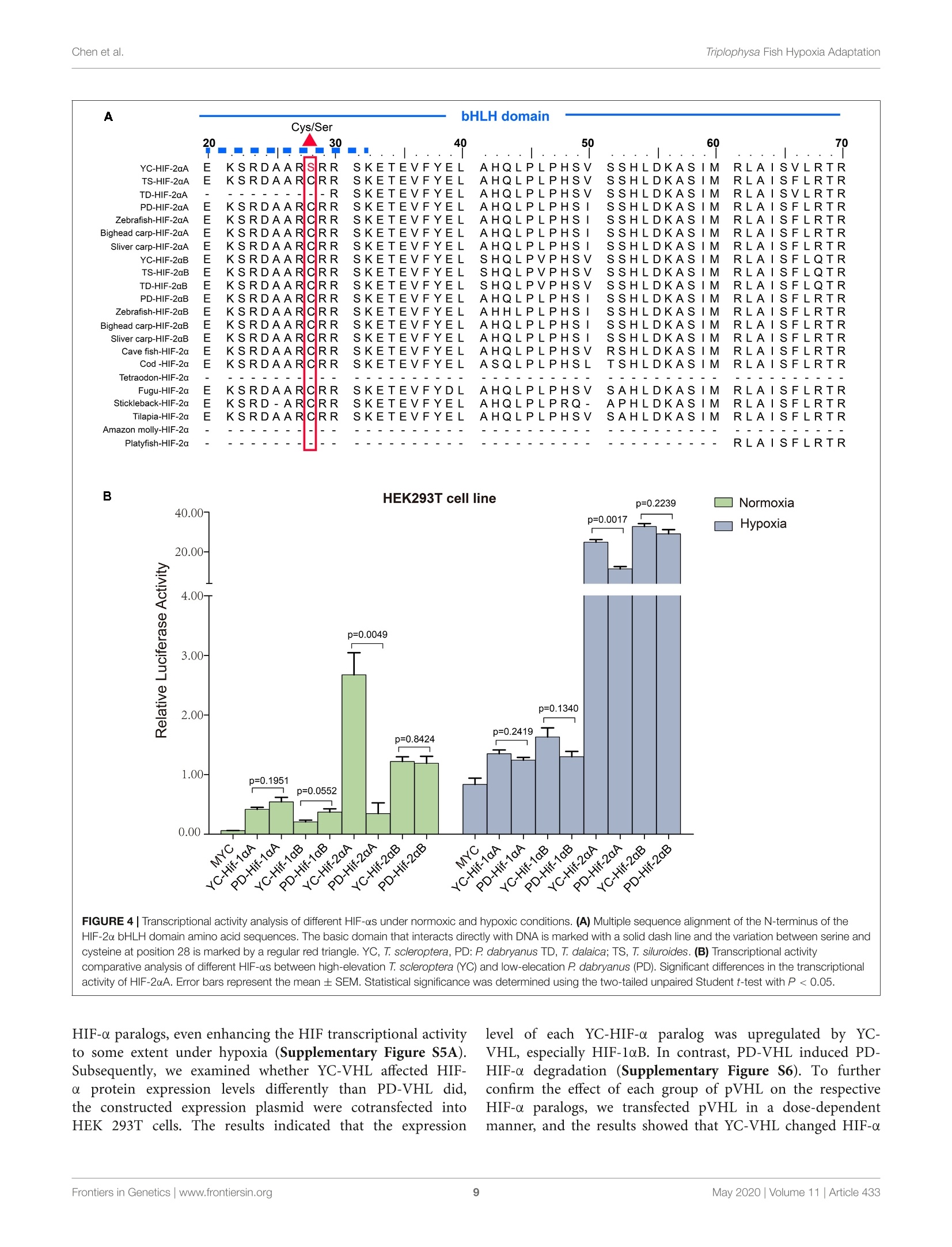
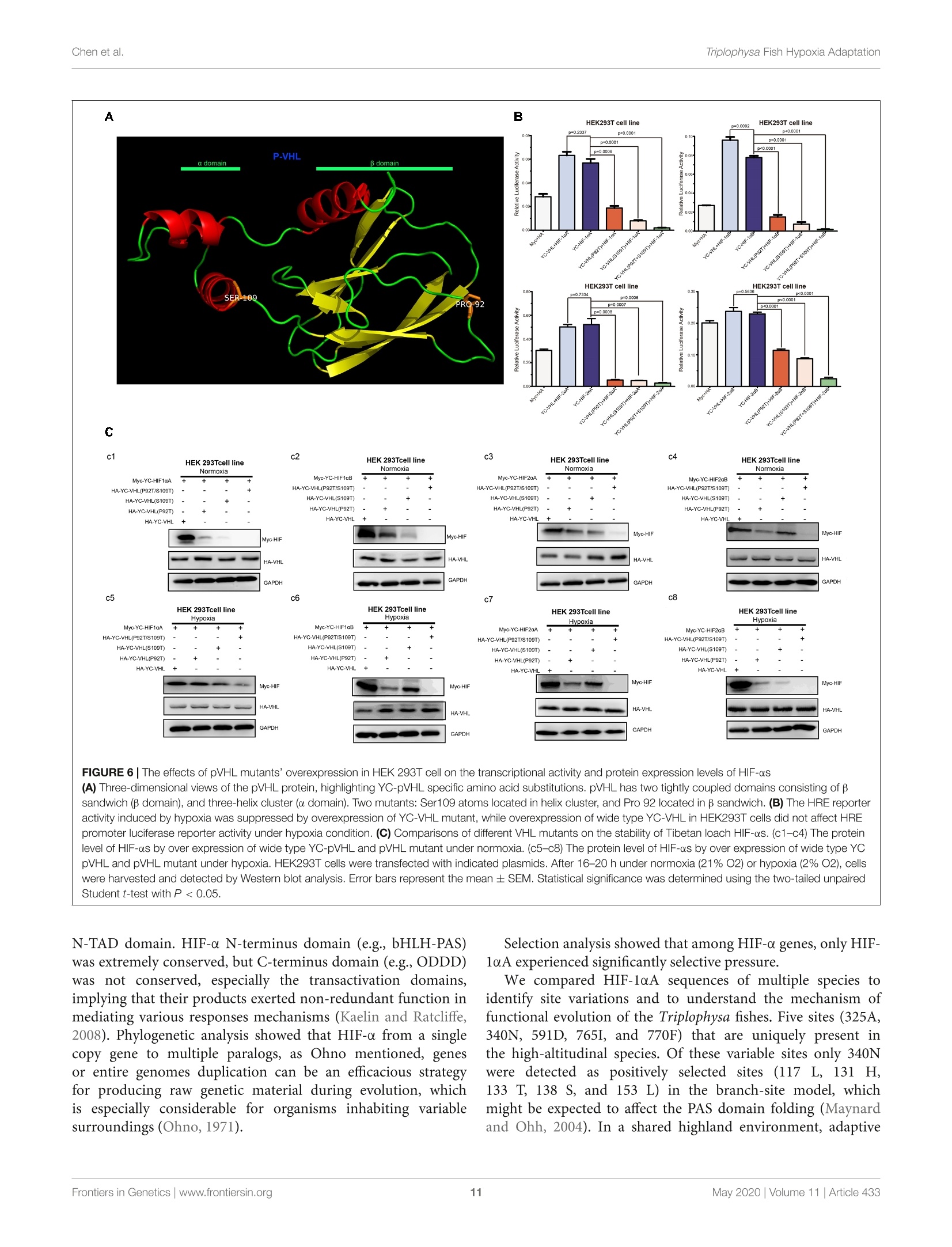
ORIGINAL RESEARCHpublished: 12 May 2020doi: 10.3389/fgene.2020.00433 Triplophysa Fish Hypoxia AdaptationChen et al. OPEN ACCESS Edited by: Tian Tang, Sun Yat-sen University, China Reviewed by:Haipeng Li,Partner Institute for Computational Biology, China Vera Maria Fonseca Almeida-Val,National Institute of Amazonian Research (INPA), Brazil *Correspondence: Liandong Yangyangliandong1987@163.comShunping Heclad@ihb.ac.cn Specialty section: This article was submitted toEvolutionary and Population Genetics,a section of the journal Frontiers in Genetics Received: 25 December 2019 Accepted: 08 April 2020 Published: 12 May 2020 Citation: Chen J, Shen Y,Wang J, Ouyang G, Kang J, Lv W, Yang L andHe S (2020) Analysis of Multiplicityof Hypoxia-Inducible Factorsin the Evolution of Triplophysa Fish(Osteichthyes: Nemacheilinae)Reveals Hypoxic EnvironmentsAdaptation to Tibetan Plateau.Front. Genet. 11:433. doi: 10.3389/fgene.2020.00433 Analysis of Multiplicity ofHypoxia-Inducible Factors in theEvolution of Triplophysa FishQ:(Osteichthyes: Nemacheilinae)Reveals Hypoxic EnvironmentsAdaptation to Tibetan Plateau Juan Chen12, Yanjun Shen12, Jing Wang3, Gang Ouyang3, Jingliang Kang12, Wenqi Lv12Liandong Yang1* and Shunping He1,4* The Key Laboratory of Aquatic Biodiversity and Conservation of Chinese Academy of Sciences, Institute of Hydrobiology,, Chinese Academy of Sciences, Wuhan, China,2University of Chinese Academy of Sciences, Beijing, China,3State KeyLaboratory of Freshwater Ecology and Biotechnology, Institute of Hydrobiology, Chinese Academy of Sciences, Wuhan,China, Center for Excellence in Animal Evolution and Genetics, Chinese Academy of Sciences, Kunming, China HIF (Hypoxia-inducible factor) gene family members function as master regulatorsof cellular and systemic oxygen homeostasis during changes in oxygen availability.Qinghai-Tibet Plateau is a natural laboratory for for long-term hypoxia and coldadaptation. In this context, T. scleroptera that is restricted to >3500 m high-altitudefreshwater rivers was selected as the model to compare with a representative speciesfrom the plain, P.dabryanus. We cloned different HIF-a and carried out a phylogeneticanalysis from invertebrates to vertebrates for identifying HIF-a genes and analyzingtheir evolutionary history. Intriguingly, the HIF-a has undergone gene duplications mightbe due to whole-genome duplication (WGD) events during evolution. PAML analysisindicated that HIF-10A was subjected to positive selection acted on specific sitesin Triplophysa lineages. To investigate the relationship between hypoxia adaptationand the regulation of HIF-a stability by pVHL in plateau and plain fish, a series ofexperiments were carried out. Comparison the luciferase transcriptional activity andprotein levels of HIF-as and the differing interactions of HIF-as with pVHL, show cleardifferences between plateau and plain fish. T. scleroptera pVHL could enhance HIF-atranscriptional activity under hypoxia, and functional validation through pVHL proteinmutagenesis showed that these mutations increased the stability of HIF-a and its heterodimerization affinity to ARNT. Our research shows that missense mutations of pVHLinduced evolutionary molecular adaptation in Triplophysa fishes living in high altitudehypoxic environments. Keywords: HIF-c, pVHL, the Tibetan Plateau, hypoxia adaption, Triplophysa fish INTRODUCTION Tibetan Plateau is commonly referred to as the “Roof of theWorld" (Zhou et al., 2006), which is the highest and largestplateau on earth. Its inhospitable environment, characterized bysevere coldness, severe hypoxia and strong ultraviolet radiation(Su and Wang, 1992; Bickler and Buck, 2007; Cheviron andBrumfield, 2012), has profound effects on animal survival andthus, it is regarded as a global biodiversity hotspot and a naturallaboratory for long-term hypoxia and cold adaptation studiesresearch (Wu and Wu, 1992; Scheinfeldt and Tishkoff, 2010;Wang et al., 2011; Qi et al., 2012; Zhang et al., 2017). Theoxygen content varies markedly daily, seasonally, and spatially(Soitamo et al., 2001).The dissolved oxygen in water decreaseswith increasing altitude, for example at 4500 m elevation, theoxygen content in the water is only 2 mg/l (Xu et al., 2016).As a representative of the endemic fishes of the Qinghai-TibetPlateau, Triplophysa scleroptera (Cypriniformes: Balitoridae:Nemacheilinae) (Herzenstein, 1888) has become an interestingmodel for studies adapted to the cold and hypoxic conditions ofits high-altitude habitat. In contrast, Paramisgurnus dabryanus iswidely distributed in the middle and lower reaches ofthe YangtzeRiver Basin. The special geographical distributions provide arich source of naturally occurring genetic variation betweenspecies, which can be used to explore the molecular mechanismof hypoxia adaptation. Previous researches primarily used high-throughput sequencing data (e.g., mitochondrial genome andtranscriptome) to explore the genetic mechanisms of highaltitude adaptations (Yang et al., 2014;Wang et al., 2015a,b,2016).However, no study has been made so far regarding the HIFpathway in Plateau fish; therefore, the impact of transcriptionalfactors and/or their target genes on the mechanisms of hypoxiaand post-hypoxia adaptation remains elusive. In this study, wefocused on the underlying molecular mechanism of hypoxiaadaptation to discover the potential genetic and physiologicalmechanism of its adaptation to high-altitude environments. The hypoxic signaling pathway is a system ofcellular signalingpathways that is very conserved from nematodes to mammals.The maintenance of oxygen homeostasis is vital to the survivalof organisms, which requires coordinated regulation of variousgenes. Hypoxia-inducible factor is the most critical factor inthis signaling system, and recent studies have shown regulationand function of HIF-a subunits in fish inhabiting differenthypoxic environments (Rahman and Thomas, 2007; Terova et al.,2008; Guan et al., 2014). HIF consists of two subunits: HIF-a and HIF- (also known as the aryl hydrocarbon receptornuclear translocator, ARNT), which belongs to the basic helix-loop-helix-PER-ARNT-SIM (bHLH-PAS) family. It's the majoractivator of downstream genes, which played a central role in thedevelopment, physiology, pathology and the adaptive responseto hypoxia (Wang et al., 1995; Semenza, 1999; Bickler andBuck, 2007). HIF-o is tightly regulated by oxygen levels, whereasHIF-B is stable (Semenza, 2012). Like other vertebrates, thereare multiple forms of HIF-as in fish. Moreover, cyprinids haveduplicate copies of all three HIF-a genes, a possible of an ancientteleost-specific third round of genome duplication (Meyer andVan de Peer, 2005; Opazo et al., 2013; Rytkonen et al., 2013; Guan et al., 2014). A key regulator of HIF-o is von Hippel Lindau(pVHL) tumor suppressor protein (Tarade et al., 2019),which hastwo domains called o and p(Stebbins et al., 1999; Cockman et al.,2000). Under normal oxygen conditions, pVHL inhibits HIF-aactivity by targeting the HIF-a subunits for polyubiquitination(Epstein et al., 2001; Jaakkola et al., 2001). Under hypoxicconditions, hypoxia reduces prolyl hydroxylase enzyme (PHD1-3) activity, stabilizing HIF-a, which joins a nuclear complex withthe constitutively expressed HIF-B and transduces the cellularresponse by binding to hypoxia-responsive elements (HREs;A/GCGTG) in the promoter regions of its target genes to activategene transcription (Wang and Semenza, 1993; Wang et al.,1995; Schofield and Ratcliffe, 2004; Kaelin and Ratcliffe, 2008;Webb et al., 2009). The relationship between HIF-as and pVHL is considered oneof the main factors that influence hypoxia signaling pathways(Bi et al., 2017). Defining the mechanisms of HIF-o regulationby pVHL will increase our understanding of the hypoxiasignaling pathway and the mechanisms of hypoxic adaptationand tolerance in highland fish. To date, the genetic mechanismsof adaptations to hypoxia have been extensively studied in theTibetan people (Beall et al., 2010; Xu et al., 2011; Bighamand Lee, 2014), Tibetan mastiff (Gou et al., 2014), Tibetangray wolf (Zhang et al., 2014), cave mammals (Shams et al.,2004), ectothermic snakes (Li et al., 2018), and diving cetaceans(Tian et al., 2016; Bi et al., 2017). However, compared tomammals, only limited amounts of genetic or other informationare available on the adaptations of fishes. Thus far, the studyon the plateau adaptability of fishes is mainly based on thetranscriptome and mitochondrial genome, investigations intochanges in evolutionary rates and to identification of the potentialgenetic bases for high-altitude adaptations (Ma et al., 2015;Wang et al., 2015a,b, 2016). Tarade et al. explored evolutionof metazoan oxygen-sensing and found the effect of pVHLon HIF-a was different (Tarade et al., 2019). However, theunderlying molecular mechanisms of hypoxia adaptation in theplateau loach remain unknown, and the important regulatoryfactors in the hypoxic pathway, including HIF-a and pVHL,have not been studied (Burga et al., 2017). In this study,to better understand the relationship between the adaptationto hypoxia and the regulation of HIF-a stability by pVHLin plateau and plain fish, we cloned and characterized fourdistinct HIF-a isoforms (HIF-1oA/Band HIF-2aA/B)andpVHLgenes to determined their expression levels and impact onphysiological regulation in T. scleroptera (highland loach) andP. dabryanus (plain loach), attempted to find differences inthe hypoxic signaling pathways between plateau fish and plainfish. This findings substantially advanced our understanding ofevolutionary biology and functional adaptation to the hypoxicecological environment of the plateau. MATERIALS AND METHODS Sample Collection Experiments involving animals in this study were conducted inaccordance with the Laboratory Animal Management Principles of China. All procedures involving the use of fish were approvedby the ethical board of the Animal Care and Use Committeeof the Institute of Hydrobiology, Chinese Academy of Sciences(protocol number IHB2017001).An adult male T. scleroptera(YC) was collected in upper reaches of the Yellow River(3600 m in elevation), where the low oxygen tension exertsunique selection pressures. Tissue samples from five majororgans including heart, liver, brain, spleen, and kidney werecollected immediately and placed in liquid nitrogen duringfield and transportation, subsequently transferred to the -80°Cfreezer for long term storage. An adult male P. dabryanus(PD) was collected in middle reaches of the Yangtze RiverBasin in Wuhan of Hubei Province (about sea level), andtissues including heart,liver, brain, spleen, and kidney were alsocollected in the same way. All of the instruments were treatedwith DEPC water. RNA Extraction, and cDNA Preparation Total RNA was, respectively, extracted from tissue samples(heart, liver, brain, spleen, and kidney) using the TRIzol reagent(Invitrogen, Karlsruhe, Germany), following the manufacturer’sinstructions. RNA degradationandcontaminationWasmonitored using 1% agarose gels. RNA concentration andthe quality were checked by spectrophotometry (optical density260/280 ratio) using a NanoDrop 2000 (Thermo Fisher Scientific,United States). RNA samples with OD260/280 ratios between1.8 and 2.1 were treated with DNase I (DNA-free, Ambion).Equal quantities of RNA were pooled from each tissue to createone mixed sample. We used 2 pg of total RNA as a templateto synthesize the first strand cDNA using reverse transcriptase(Promega) in the presence of RNase inhibitor (Invitrogen) witha cocktail of oligoT and dNTP (TaKaRa) and rapid amplificationof cDNA ends (RACE) using a Clontech SMARTerTM RACEcDNA Amplification Kit. Aliquots of 1 ul of undiluted cDNAwere used per PCR. Molecular Cloning and PlasmidConstructs We searched for these desired genes (HIF-as and pVHL)in the available transcriptome sequences. The genome-widesurvey was based on TBLASTN searches in the separatetranscriptome assemblies: T.scleroptera (PRJNA280009), andP. dabryanus (PRJNA266739). ORF finder on NCBI was usedto predict the amino acid sequences of HIF-as and pVHL.SMART (Letunic et al., 2015) and UniProt were utilizedto check protein domains. Based on our transcriptome data,we designed a pair of primers by using Primer Version 5.00(Primer Biosoft International, Palo Alto, CA, United States),to obtain the true sequence of the gene by PCR technology.Primers for amplifying are listed in Additional file 1: TableS1. Subsequently, the PCR products were recombined withpMD18-T vector (TaKaRa), sequenced using an ABI3730XLsequencer, and assembled using SeqMan in DNASTAR Lasergenev7.1 software (DNASTAR Inc.). Mutagenesis of the pVHLsequence (P92 T, S109 T, and P92 T+S109 T) was achieved through site-directed mutagenesis andd verified by Sangersequencing. Briefly, the complementary oligo deoxyribo-nucleotide primers were designed at the mutation sites, andstandard PCR cloning techniques wereuused to generatefragments that included the mutations. 工Thee correctandintact full-length pVHL, HIF-1aA/B and HIF-2cA/B fromT. scleroptera and P. dabryanus were isolated from one of theTA clones using newly designed gene-specific primers to the5 and 3' ends with restriction enzyme sites (Additional file2: Supplementary Table S2), and ligated into the expressionvector PCMV-HA.(CloneTech), and the PCMV-Myc vector(CloneTech)..All recombinedexpressionplasmidswereverified by Wuhan Tianyi Huiyuan Bioscience & TechnologyCorporation (Wuhan, China). Sequence and Evolutionary Analysis For phylogenic analysis, sequences of the HIF-a (HIF-1oA/B,HIF-2oA/B) and pVHL were obtained from GenBank’ andEnsemble’. In total, 70 homologous sequences for HIF-a wereretrieved and sequence identity was performed using onlinetools. Sequence alignments were performed using MUSCLE(Edgar, 2004). Additionally, jModeltest 2.1.10 (Posada, 2008)was used to choose the best models for nucleotide substitution.Finally, a phylogenetic tree was constructed by MrBayes-3.2.2(Huelsenbeck and Ronquist, 2001) with 20,000,000 iterations.Toinvestigate whether HIF-a and pVHL genes have undergonestatistically significant differences in selection pressures, weemployed branch-site model in the CodeML program (PAMLV4.8) (Yang, 2007) to detect the positive selection that mayhave affected specific sites along a specific branch. In thebranch-site model (model = 2, Nsites =2); the neutral model,which constrains a class of sites to ω = 1(fix omega = 1,omega =1); and the selection model that allows a class ofcodons in the foreground branch to have ω>1 (fix omega=0,omega =1.5). Likelihood ratio tests were used to comparethese nested models with a x2 distribution was used to identifypositively selected codons. Positive selected sites were detectedby the Bayes empirical Bayes method (Yang et al., 2005).In addition, the PROVEAN (Choi et al., 2012) was used topredict whether an amino acid substitution or indel has animpact on the biological function of a specific site. Variantsmost likely alter protein function. The PROVEAN score wascalculated for each variant; the more negative the score, the morelikely a given variant alters the protein function. Furthermore,the 3D structures of the pVHL gene were constructed byusing Swiss-Model (Bordoli et al., 2009), and the subsequentpVHL molecular visualization performed by using PyMOL(Schrodinger, 2010). Cell Culture Human embryonic kidney (HEK) 293 T cells were usedbecause they possess high transfection efficiency. HEK 293 Tcells were cultured in Dulbecco’s modified Eagle’s medium(DMEM, Hyclone)Ssupplementedwith10%fetalbovine ( ht tp://www.ncbi.nlm.nih.gov/genbank/ ) serum (FBS, Hyclone) and 1% penicillin and streptomycinat 37C in aa5% CO2 humidified incubators (Thermo).Hypoxia treatment was conducted in a hypoxic chamberwith 2% oxygen concentration by using an incubator withO2 control filled with 5% CO2 and balanced with N2(NBSGalaxy 48R). Cells were plated in six-well plates or twentyfour -well plates at 50-70% confluence 1 day before thetransient transfection. Western Blot Analysis For protein expression assays, HEK 293 T cells were seeded in6-well plates for 12 h before transfection, and then transfected withthe indicated amounts of plasmids using VigoFect (Vigorous)reagent. Transfected cells were incubated in fresh medium anddivided into two groups and cultured in normoxic or hypoxicchambers (2%O2) (Ruskinn INVIVO2I-400) for 18 h. After twoconsecutive washing steps with PBS, the cells were harvested inRIPA buffer (Beyotime) with 1% PMSF (Beyotime), 1% Na3VO4,and a phosphatase inhibitor cocktail (Thermo Scientific) wasadded. All operations are done on ice for 50 min, and celldebris was removed by centrifugation at 13,000 rpm for 15 min.Then the lysate was mixed with 5× protein loading buffer(Beyotime), and boiled for 10 min. The antibodies were usedas follows: anti-c-Myc antibody (9E10, Santa Cruz), anti-HAantibody (Covance), and anti-GAPDH antibody (60004-1-Ig).HRP labeled goat anti-mouse IgG (H + L, Beyotime) wasused as the secondary antibody. We used a Fujifilm LAS4000luminescent image analyzer to photograph the blots. ImageJsoftware was used to evaluate quantify protein levels basedon density of the protein bands. For the analysis of thedegradation rate of HIF-as in the highland loach and plainfish, Myc-tagged HIF-as was transfected into the HEK293Tcells in the presence of cycloheximide (CHX, 10 ug/ml; Sigma,Munich, Germany), which was used to block the synthesis ofendogenous HIF-as. Transfections and Luciferase ReporterAssays Cells were seeded in 24-well plates for 12 h under normoxia(21%02))before transfection and then were transfectedwithh the indicated amounts of plasmidsuusingVigoFect(Vigorous) together with the hypoxia response element luciferasereporter (HRE-Luc.) following the instructions recommendedin the manufacturer’s protocol. pRL-Renillai vwasusedasan internal control. Luciferase activity was measured 24 hafter transfection using the Dual-luciferase Reporter AssaySystem (Promega). Luciferase activity data was normalized toRenilla luciferase. All experiments were performed in threeindependent operations. Statistical Analysis Luciferase assays data were normalized to Renilla luciferase andpresented as mean ± SE.M. of three independent experimentsperformed in triplicate. We compared mean relative luciferaseactivity by using unpaired Student's t tests in GraphPad Prism 5(GraphPad Software, La Jolla, CA, United States). Characterization of HIF-0 Duplicationsand pVHL Gene The sequences of the HIF-a genes (HIF-1A/B and HIF-2A/B)and pVHL were obtained here for high -land Triplophysa fish andfor fishes living at low altitudes (P. dabryanus). Bioinformaticsanalysis showed that all HIF-a/pVHL genes contained completeORF regions, and the biochemical and physical parameters arelisted in Additional file 3: Supplementary Table S3. A computeranalysis indicated that the T. scleroptera ORF of the HIF-1aA, HIF-1oB, HIF-2oA and HIF-2oB cDNA encodes for aputative protein of 686,776, 843, 817 amino acids, respectively,and shares ~40-56% sequence similarity with the mammalianHIF-as, and high shared identity with other Triplophysa fishes.On the other hand, analysis of the pVHL cDNA showedthat it encodes a putative protein of 171 amino acids, whichshares >80% sequence similarity with PD pVHL (Additionalfile 4: Supplementary Table S4). Both HIF-1oA/B and HIF-2A/B deduced proteins contain the characteristic domainssimilar to those of their mammalian counterparts: bHLHdomain, PAS-A and PAS-B domains, PAC motif, an oxygen-dependent degradation domain (ODDD), and two TADs domain(Supplementary Figures S1, S2). Furthermore, the HIF-asubunits have N-NLS and C-NLS (nuclear localization signals),respectively. HIF-1aA and HIF-1oB were about 60% identical,while HIF-2A and HIF-2aB were 62.23% identical. HIF-1ois structurally similar to HIF-2a; these two subunits share43% amino acid sequence identity and are mainly differed inthe N-TAD domain.We found that the N-terminal domains(bHLH, PAS-A/B, and C-TAD) were highly conserved, while theC-terminal domains especially ODDD, which responsible for theoxygen-dependent degradation of the protein (Ivan et al., 2001;Rytkonen et al., 2007), had lower identity values. This suggestingthat they may be differences in their sensitivity to hypoxia.The hydroxylation targets, the proline and the asparagine whichare located in the ODDD, C-TAD domain, respectively, areconserved, despite the highly variable sequences among them.In addition, we found some shared amino acid replacementsin the HIF-a paralogs of the Tibetan loach, which is locatedin important domains (Figure 1, Supplementary Figure S3).At the same time, we found that some basic amino acidsthat interact with DNA, namely, K19, K21, R27, R29, R30,D55, and K56 (HIF-1a), appear to be conserved across species,indicating integrity function with DNA binding (SupplementaryFigures S1, S2). Sequence analysis indicated that the deduced amino acidsequence of pVHL also included characteristic a/p domainsimilar to mammalian counterparts. Some amino acid siteswere uniquely found in the high-altitude loach, which differedfrom lower-altitude pVHL (Supplementary Figure S4).It iswell known that site differences often determine a differencein function. Whether the difference between these sites affectsthe function of the gene, should be explored.. We usedPROVEAN to calculate the possible impact of each variant,and the default threshold was-2.5. When the score equal FIGURE 1|Phylogenetic tree of the HIF-a genes. The maximum likelihood method was used to reconstruct the tree that was rooted with invertebrate HIF-asequences. Species are listed along with their phylums or classes. to or less than -2.5 were considered“deleterious, while thescore greater than -2.5 were considered “neutral."The morenegative the score, the more likely a given variant is toalter the protein function. Most variants were predicted tobe neutral; however, the proline-to-threonine replacement atposition 92, which is located in the B domain (the HIF-binding domain), was predicted to be deleterious (Additionalfile 5: Supplementary Table S5). In addition, another aminoreplacement uniquely exists in YC-pVHL at position 109 in thea domain (Supplementary Figure S4B), where ElonginC binds in association with ElonginB to recruit the pVHL-ElonginB-ElonginC complex to Cul2-Rbx1, which forms a ubiquitin-protein ligase. Phylogenetic Analysis and SelectivePressure Analysis The phylogenetic analysis of the HIF-as acrosssd(ifferentorganisms or species with MrBayes recovered the tree shownin Figure 1. Regarding early eukaryotic evolutionary history, only one HIF-a has been discovered in the genomes of theexamined invertebrates, such as Caenorhabditis elegans. However,where amphioxus at the most basal of chordate subphylum has asingle HIF-a gene, vertebrates often have two or more paralogsderived from two whole-genome duplication events, suggestingthat amphioxus were the ancient origin of the HIF-a family.The additional two paralogs (HIF-1oA/B and HIF-2oA/B) werepresent in cyprinids most likely a result from the teleost-specificwhole-genome duplication (Rytkonen et al., 2013) (Additionalfile 6: Supplementary Table S6). The topology of HIF-a memberswere largely consistent with the results based on mitogenome andtranscriptome data in previous studies. The phylogenetic analysisstrongly support three clades (Figure 1). The deduced amino acidsequences of HIF-3o and HIF-4o forming a separate group, as asister group of HIF-10. To better understand whether the hypoxia-related gene wassubjected to natural selection during plateau adaptation, weestimate the frequency on the coding sequences. We used theone ratio model in the CodeML for the phylogenetic analysiswith maximum-likelihood (ML) software (PAML V4.7). The ωvalues of these hypoxia-related gene ranged from 0.15 to 0.17an werree significantly <1 (Additional file 7: Supplementary stabiliTable S7), suggesting under strong purifying selection processin the evolution. To investigate whether positive selection acted on specificsites in Triplophysa lineages, we used branch-site models inPAML. In all the HIF-a paralogs, we found that only HIF-loA(pp<5.933e-07) subjected to positive selection thatacted on specific sites. By using the BEB (Bayes empiricalBayes) method, there were 16 amino sites subjected to positiveselection in HIF-1aAwitlh a posterior probability greaterthan 0.5 (Additional file 8: Supplementary Table S8). Someof the Triplophysa lineage-specific nonsynonymous mutationswere identified, including 325A, 340N, 591D,765I, and 770F(Figure 2A). The 340th asparagine had a high posteriorprobability of 0.989, which was significant at the 5% confidenceinterval. The highland loaches possess a unique amino acidasparagine, in an important PAC domain; however, serine,not arginine,was found in all representative vertebrates,and may be important for PAS domain folding (Maynardand Ohh, 2004). For pVHL, the branch-site model was alsocarried out to detect positive selected sites along a specificbranch. When we set all Triplophysa fishes as the foregroundbranches, did not find any positive selection signal. To furtherexplore the potential difference, a clade of T. scleropterawas set as the foreground branches, and we found someunique, likely positive selection sites (19A, 61T, 98A, and109S in T. scleroptera, which all located in important domains(Supplementary Figure S4B). HIF-o Paralog Proteins Have DivergentExpression As two major regulatory factors of the hypoxia signalingpathway, HIF-1a and HIF-2o have garnered long-term interestin distinguishing their roles, and related research have shownITF-1nthat both HIF-1a and HIF-2o participate in hypoxia-dependent gene regulation (Raval et al., 2005). The discovery of differencesbetween HIF-a isoforms could advance our understanding ofhigh-altitude hypoxic stress response mechanisms. To test thepotential functional differences ofthe HIF-as protein in highlandloach compared with the lowland species (SupplementaryFigure 3A), we compared of these protein level under hypoxic ornormoxic conditions.Western blot analysis displayed that all thefish HIF-as could be expressed in HEK 293T cells and the proteinexpression levels of each HIF-a subunits are different fromeach other. Compared to the other HIF-a subunits, HIF-1aBprotein expression level is the highest neither hypoxic conditionsnor normoxic conditions in high-elevation fish. Nevertheless,HIF-1aA is the most highly expressed isoform in plain fish(Figure 2B). Generally, whether under normoxia or hypoxicconditions, HIF-1a was greater protein expression level thanHIF-2o, which implying that HIF-1a may be acted as animportant regulator of fish in response to the environmentalhypoxia (Figure 2B). Compared to those of highland fish, theincrease in the levels of protein expression of plain fish wasrare. Subsequently, to investigate whether there is a differencein hypoxia-induced regulation of HIF-1as and HIF-2as proteinstability between highland and plain fish, CHX was used to blocknew protein synthesis. We found that HIF-a protein from theP. dabryanus (PD) were degraded at a higher rate than those fromthe T. scleroptera (YC) in response to hypoxia (Figure 3).Thedegradation rate of HIF-a was slower in YC, suggesting long-term highland exposure enhanced gene expression function foradaptation to hypoxic environments. These results promptedus to ponder whether other genes in the signaling pathwaymight invoke differences between highland fishand low-elevation fish. HIF-2A More Efficiently Induced TargetGene Expression Compared With OtherHIF-c Isoforms To assess the transcriptional activity of different HIF-a paralogsin highland loach and plain loach, Myc-tagged HIF-a plasmidwere cotransfected withHREand pRL-Renillaaluciferasereporters into HEK 293T cells. Our luciferase reporter assay indicated all the HIF-a paralogscould upregulate the luciferase activity of the HRE reporter genethrough dimerization with human HIF-B. Different HIF-a formsshowed different levels of transcriptional activity. In general,hypoxia could enhance the transcriptional activity of HIF-as.Intriguingly, in comparison with plain loach, HIF-2aA fromthe high-land loach exhibited stronger effects on transcriptionwhether under normoxic or hypoxic conditions (p <0.05).However, there were no significant differences among the otherHIF-aparalogs (Figure 4B). Subsequently, through multiplecomparative analysis of amino acid sequences for HIF-2aA,we found that the only difference in the DNA-binding basicregions was that the cysteine found in all other representativefishes correlated with serine 28 in T. scleroptera (Figure 4A).As Okuno mentioned, this nonsynonymous mutation makescysteines mutate serines, increasing the transcriptional activity byenhance affinity with DNA-binding complex(Okuno et al., 1993). YC-VHL Stabilizes the HIF- Potein andEnhances the Transcriptional Activity ofHIF-c Genes Hypoxia.1inducible: factor (HIF) -mediated transcriptionalactivation is the most critical signaling pathway for cells tosense hypoxia, in this process, pVHL is crucial to the stabilityof HIF-a. In light of the well-understood function of HIFa inhypoxia signaling and hypoxia adaptation for fishes living athigh elevation, we next studied whether T. scleroptera pVHL is unique compared to that of species in the plain habitat. Initially,we examined whether pVHL could affect HIF-a transactivity indifferent ways, by cotransfecting HA-taggedpVHL together withMyc-tagged HIF-a, HRE-luciferase reporter and pRL-Renillaluciferase reporters into HEK 293T cells. Overexpression ofPD-VHL significantly inhibited the transactivity of the HIF-aparalogs (Supplementary Figure S5B) under either normoxicor hypoxic condition. In contrast, YC-VHL could considerablysuppress the transactivity of HIF-a paralogs under normoxicconditions, whereas YC-VHL did not inhibit the transactivity of HIF-a paralogs, even enhancing the HIF transcriptional activityto some extent under hypoxia (Supplementary Figure S5A).Subsequently, we examined whether YC-VHL affected HIF-a protein expression levels differently than PD-VHL did,the constructed expression plasmid were cotransfected intoHEK 293T cells. The results indicated that the expression level of each YC-HIF-a paralog was upregulated by YC-VHL, especially HIF-1oB. In contrast, PD-VHL induced PD-HIF-a degradation (Supplementary Figure S6). To furtherconfirm the effect of each group of pVHL on the respectiveHIF-a paralogs, we transfected pVHL in a dose-dependentmanner, and the results showed that YC-VHL changed HIF-0 FIGURE 5|The effects of different concentration gradients pVHL on the protein expression of HIF-as under hypoxia. (A) The effect of YC-VHLon Tibetan loach (YC)HIF-a isoforms (HIF-1αA/B and HIF-2aA/B) under hypoxic conditions. (B) The effect of PD-VHL on plain loach (PD) HIF-a isoforms under hypoxicconditions.HEK293T cells were co-transfected with equal amounts of Myc-tagged HIF-as expression vector along with increasing amounts of HA-tagged pVHLexpression vector under hypoxia. stability to a negligible degree, and upgraded HIF-a proteinexpression levels (e.g., HIF-1oB). However,PD-VHL promotedP.dabryanus HIF-a degradation in a dose-dependent manner(Figure 5). Through the further inspection of the amino acidsequences for pVHL coupled with selection analysis and 3Dstructure, we obtained three mutants of YC-VHL that mimicthe amino acids of PD-VHL; specifically, Pro at position 92was mutated to Thr (YC-VHL-P92T), Ser at position 109 wasmutated to Thr (YC-VHL-S109T), and the two sites weresimultaneously mutated (YC-VHL-P92T + S109T). Luciferasereporter assays revealed that YC-VHL site mutants repressedthe HRE reporter activity activated by HIF-a overexpression,while the wide type YC-VHL did not significantly affectHRE-reporter activity under hypoxic conditions (Figure 6B).Furthermore, this result confirmed by western blot assays.YC-VHL mutants could induce HIF-a degradationi underhypoxia or normoxia to an extent similar to that of PD-VHL (Figure 6B). These results not only show that theinduction of HIF-a degradation owing to the uniqueness of PD-VHL, but also indicate that YC-VHL may uniquely modulatehypoxia signaling, thus promoting hypoxia tolerance by meansof stabilizing HIF-a proteins expression and strengtheningtranscriptional activity. DISCUSSION Animals living on the plateau or in caves, such as birds, mammalsand fishes, have been exposed to a low-oxygen environment for along time (Ramirez et al., 2007). The primary research on hypoxiaadaptation mainly focused on endothermic vertebrates. However,studies performed on poikilothermic animals is relatively scarce.The fish fauna of the Qinghai-Tibet Plateau is mainly composedofthree groups, namely, the Schizothoracine fish, Glyptosternoids,and Triplophysa, which are considered excellent fish models for studying high-altitude adaptation (Kang et al., 2017). They are allwell adapted to the harsh ecological environment of the QinghaiTibet Plateau, characterized by the cold, hypoxia and strong UV.HIF-a and pVHL, as the key factors in hypoxia signaling pathway,dominating the main signal regulation network governingcells in response to hypoxia (Graham and McCracken, 2019),may diverge between high-altitude and low-elevation species.Although previous studies identified HIF-as in highland fishes(Cao et al., 2008; Guan et al.,2014), however, all previous studiesof fishes focused only on HIF-a sequence analysis or simplefunctional analysis of individual genes. The present study basedon the mechanisms of regulation of HIF-a subunits by pVHLwill increase our understanding of the hypoxia signaling pathwayalong with help us to understand the mechanisms of high-altitudeadaptation of the Triplophysa fishes to the aquatic fields of theQinghai-Tibet Plateau. Environmental heterogeneity affects the adaptive evolutionarytrajectory ofspecies. Oxygen capacity often affectss thephysiological, biochemical and life cycle activities of fishes,and also has a profound impact on both the phenotypesand geographical distributions of living beings. For instance,Carassius carassius can survive several months in a cold andhypoxic environment (Nilsson and Ostlund-Nilsson, 2004). Incontrast, the Wuchang bream (Megalobrama amblycephala)is a hypoxia-sensitive species, hence, even transitory hypoxiatreatment at room temperature is fatal (Ouyang et al., 2001). Thegenus Triplophysa endemic to the Tibetan plateau and experienceextreme environmental conditions. Therefore, these speciespresent an ideal opportunity to study the hypoxia adaptationof fishes. With comparative analysis of amino acid sequencesof multiple species, we found that each HIF-a contained themain domains and the key residues (proline residues) found intheir mammalian counterparts (Prabhakar and Semenza, 2012).HIF-1o is structurally similar to HIF-2o; these two subunits share43% amino acid sequence identity and are mainly differed in the FIGURE 6| The effects of pVHL mutants’overexpression in HEK 293T cell on the transcriptional activity and protein expression levels of HIF-as(A) Three-dimensional views of the pVHL protein, highlighting YC-pVHL specific amino acid substitutions. pVHL has two tightly coupled domains consisting of βsandwich (B domain), and three-helix cluster (a domain). Two mutants: Ser109 atoms located in helix cluster, and Pro 92 located in p sandwich.(B) The HRE reporteractivity induced by hypoxia was suppressed by overexpression of YC-VHL mutant, while overexpression of wide type YC-VHL in HEK293T cells did not affect HREpromoter luciferase reporter activity under hypoxia condition. (C) Comparisons of different VHL mutants on the stability of Tibetan loach HIF-as. (c1-c4) The proteinlevel of HIF-as by over expression of wide type YC-pVHL and pVHL mutant under normoxia. (c5-c8) The protein level of HIF-as by over expression of wide type YCpVHL and pVHL mutant under hypoxia. HEK293T cells were transfected with indicated plasmids. After 16-20 h under normoxia (21%O2) or hypoxia (2%O2), cellswere harvested and detected by Western blot analysis. Error bars represent the mean ± SEM. Statistical significance was determined using the two-tailed unpairedStudent t-test with P <0.05. N-TAD domain. HIF-a N-terminus domain(e.g., bHLH-PAS)was extremely conserved, but C-terminus domain (e.g., ODDD)was not conserved, especially the transactivation domains,implying that their products exerted non-redundant function inmediating various responses mechanisms (Kaelin and Ratcliffe,2008). Phylogenetic analysis showed that HIF-a from a singlecopy gene to multiple paralogs, as Ohno mentioned, genesor entire genomes duplication can be an efficacious strategyfor producing raw genetic material during evolution, whichis especially considerable for organisms inhabiting variablesurroundings (Ohno, 1971). Selection analysis showed that among HIF-a genes, only HIF-1aA experienced significantly selective pressure. We compared HIF-1oA sequences of multiple species toidentify site variations and to understand the mechanism offunctional evolution of the Triplophysa fishes. Five sites (325A,340N, 591D,765I, and 770F) that are uniquely present inthe high-altitudinal species. Of these variable sites only 340Nwere detected aspositively selected sites (117 L, 131 EH,133 T, 138 S, and 153 L) in the branch-site model, whichmight be expected to affect the PAS domain folding (Maynardand Ohh, 2004). In a shared highland environment, adaptive selection for chronic hypoxia is a vital unit of the evolutionfor aboriginal species. Previous studies, including ectothermicsnakes (Li et al., 2018), Tibetans (Beall et al., 2010), Tibetanhorses (Liu et al., 2019), and Tibetan mastiff (Miao et al.,2016), have identified EPAS1, also named HIF-2a shows thestrongest semaphore for positive selection. Given the results ofour study together with previous study of schizothoracine fish(Guan et al., 2014), significant selective pressure was detectedin HIF-1a. It appears that the fishes have developed distinctmechanisms to adapt the unfavorable ecological environment ofthe Tibetan Plateau. FunctionalanalysiswaSperformedlLby transfection ofHIF-as and pVHL into HEK 293T cells. Different HIF-asperformed different transcriptional activities. Luciferase analysesin the current study revealed that HIF-2oA isoforms moreefficiently inducedl targetgene expression. Divergence oftranscriptional activity usually been related to differencesin N-terminal transactivation domains between HIF-1a andHIF-2a, which imply that intrinsic differences of HIF-aisoforms coding sequences might facilitate isoform-particularfunction (Koh and Powis, 2012). Generally, during chronichypoxic exposure, HIF-2a variants plays an important majorrole in driving the hypoxic reaction (Hu et al., 2003; Kohand Powis, 2012). In comparison with plain loach, Tibetanloach exhibit dramatic increases in the transcription activityof HIF-2oA. The specific nonsynonymous mutation of theconserved cysteines to serines at position 28, yielded a strongeraffinity DNA-binding compound, subsequently strengthenedcell transforming ability (Lando et al., 2000; Rytkonen et al.,2007). Although the HIF-2a transcriptional activity is higherthan HIF-1a, nevertheless the protein expression level appearedantithetictendency,. suggestingHIF-aproteinlevelsarerelated toposttranscriptional or posttranslational changes(Kietzmann et al., 2016).This result is similar to that foundfor schizothoracine fish, which also located on the TibetanPlateau (Guan et al., 2014). By Comparing of protein leveldegradation rates,wefound that HIF-a proteins fromP. dabryanus (PD) were degraded faster than those of theT. scleroptera (YC) in response to hypoxia. T. scleroptera HIF-asare more stable than those of low-elevation loach, leading toaccumulation and are thought to enhance cell proliferation,migration, and. survivalfor chronicic hhypoxiaadaptation(Stantic et al., 2018). HIF-a and pVHL, as the key factors in the hypoxia signalingpathway, might diverge between organisms in high-altitudeadaptation and lowland species. After aligning the amino acidsequences of pVHL from different species, we found that someunique amino acid sites were detected as possible positivelyselected sites (19 A, 61 T, 98 A, 109 S), restricted to thehighest-altitudinal species T. scleroptera. Notably, proline-to-threonine substitution at position 92, which is located inthe B domain might affects protein function as predicted bythe PROVEAN. Exploring regulatory mechanisms of HIF-asby pVHL will improve our understanding of the hypoxiasignaling pathway and plateau adaptation. To test the potentialfunctional effects of pVHL in regulating HIF stability, wecompared regulatory mechanisms in Tibetan Plateau compared with the lowland species. T. scleroptera wild-type pVHL (YC-VHL) did not influence HER-reporter activity under hypoxia.However P. dabryanus pVHL (PD-VHL) could decline thetranscriptional activity of HIF-as. Using YC-VHL mutants(P92T, S109T) to mimic amino acids of low elevation allele,our results showed that the pVHL mutant could suppress thetranscriptional activity of YC-HIF-as under hypoxia, suggestingthat YC-VHL may uniquely modulate hypoxia signaling. Itisworth noting that P/S are all non-polar amino acidswhereass T is a polar amino acid and this mutation mightleads to protein conformational diversification. The uniquefeature of YC-VHL may contribute to the specific high-altitude adaptation. Furthermore, we revealed that YC-VHLfacilitates hypoxia tolerance by stabilizing HIF-a proteins,whereashPD-VHL could induce HIF-a degradation underhypoxia or normoxia. Notably, Our site-directed mutagenesisexperiment further demonstrated that YC-VHL can stabilizeHIF-a proteins. These phenomena are consistent with aquaticcetaceans unique feature of pVHL to hypoxia adaptation,which possess divergent diving capabilities with commensuratedifferences in oxygenation (Bi et al., 2017). Even though wehave boldly explained this phenomenon with some experimentalevidences, we cannot preclude the existence of other signalpathways to mediate HIF-a function in hypoxia tolerance(Webb et al., 2009). HIF-a regulation is complicated infishes and might be shaped by multiple physiological andenvironmental factors. Further research is indispensable toinvestigate the structural and functional differences of HIF-a subunits. DATA AVAILABILITY STATEMENT The sequencing data have been deposited into the NationalCenter for Biotechnology Information (GenBank: MN990446-MN990455). ETHICS STATEMENT Experiments involving animals in this study were conducted inaccordance with the Laboratory Animal Management Principlesof China. All experimental protocols were approved by theEthics Committee of the Institute of Hydrobiology, ChineseAcademy of Sciences. AUTHOR CONTRIBUTIONS SH and LY led the project. JC performed the experiments,analyzed the data, and wrote and revised the manuscript. YS,JW, GO, JK, and WL participated in fish sampling and providedadvices in experiments. FUNDING This work was supported by grants from the National NaturalScience Foundation of China (91731301), from the Strategic Priority Research Program (XDB13020100) to SH, and fromthe National Natural Science Foundation of China (31601858and 31972866) to LY. This research was supported by theWuhan Branch, Supercomputing Center, Chinese Academy ofSciences,China. ACKNOWLEDGMENTS We thank all the members from lab for help. We also thank GO,Sijia Fan, and Chenxi Xu for help and generous gift of reagents. SUPPLEMENTARY MATERIAL The Supplementary Material for this article can be foundonlineeat: https://www.frontiersin.org/articles/10.3389/fgene.2020.00433/full#supplementary-material FIGURE S1 | Multiple sequence alignment of the deduced HIF-1a proteinsequences from human, zebrafish, T. scleroptera (YC), P. dabryanus (PD). Dashesindicate the gaps inserted to facilitate alignment. The main domain of HIF-1aA/B ismarked with a solid line box or an overline. The two conserved proline residueswithin the ODDD domain and the asparagine residue in C-TAD are indicated bythe red arrows. The gray boxes represent two conserved proline hydroxylationmotif LxxLAP areas. The DNA-interacting basic amino acids K19, K21, R27, R29,R30, D55, and K56 are highlighted in green. FIGURE S2|Multiple sequence alignment of the deduced HIF-2a proteinsequences from human, zebrafish, T. scleroptera (YC), P. dabryanus (PD). Dashesindicate the gaps inserted to facilitate alignment. The main domain of HIF-1aA/B ismarked with a solid line box or an overline. The two conserved proline residueswithin the ODDD domain and the asparagine residue in C-TAD are indicated bythe red arrows. The gray boxes represent two conserved proline hydroxylationmotif LxxLAP areas. The DNA-interacting basic amino acids K16, K18,R24, R26,R27, D52, and K53 are highlighted in green. FIGURE S3|Domain structure of hypoxia-inducible factor subunits. (A) Domainstructure of HIF-1B. (B) Domain structure of HIF-2aA. (C) Domain structure ofHIF-2B. The following domains are shown: basic helix-loop-helix domain (bHLH),Per-Arnt-Sim homology domain (PAS-A/B), a PAS-associated COOH-terminal(PAC) motif, O2-dependent degradation domain (ODDD), NH2- and REFERENCES ( B eall, C . M ., Cavalleri, G. L ., D e ng, L., Elston, R. C. , Gao , Y., Kn i ght, J., et al.(2010) . Natural s election on EPAS1 ( HIF2alpha) a s sociated w ith low hemoglobinconcentration in Ti b etan highlanders. P r oc. Natl. A cad. Sci. U.S.A. 107, 11459- 11464. doi: 10.1073/pnas.1002443107 ) ( Bi,J., Hu, B ., Wang, J., Liu, X ., Z heng, J . , W ang, D., et al. (2017). B e luga whalepVHL e n hances HIF-2alpha ac t ivity via i nducing HIF-2alpha proteasomaldegradation under h y poxia. O n cotarget 8, 42272-42287. doi: 1 0 .18632/ oncotarget.15038 ) ( Bickler, P. E., and Buck,L.T. (2007). Hypoxia tolerance in r eptiles, amphibians, and f ishes: life with variable oxygen a v ailability. Annu. Rev. Physiol. 69 , 145 - 170. doi:10. 1 146/annurev.physiol.69.031905.162529 ) ( Bigham, A. W., a n d Lee, F . S. ( 2014). H uman hi g h-altitude adaptation: forwardgenetics meets the HIF p athway. Genes D e v. 28, 2189-2204. doi: 1 0 .1101/gad.250167.114 ) ( Bordoli, L ., Kiefer, F. , Arnold, K., Benkert, P., Battey, J ., an d Schwede, T . (2009).Protein structure homology modeling usin g SWISS-MODEL w o rkspace. Nat. P rotoc. 4 , 1 -13. doi: 10.1038/nprot.2008.197 ) ( B urga, A., Wang, W., Ben-David, E., Wolf, P. C., Ramey, A. M., Verdugo, C., et al.(2017). A genetic signature o f t h e e v olution of loss of flight in the Galapagos cormorant. Science 356:eaal3345. doi: 10.11 2 6/science.aal3345 ) COOH-terminal transactivation domains (TAD-N and TAD-C). Percentagerepresents the similarity ratio of the domain between highland loach (YC:T. scleroptera) and plain loach (PD: P. dabryanus). Amino acid replacements inTriplophysa compared with all low-elevation species are shown in red. TD,T. dalaica; TS, T. siluroides. FIGURE S4|Sequence and domain analysis of pVHL (A) Multiple sequencealignment of the deduced pVHL protein sequences. The red underline indicates βdomain, the blue dotted line represents the a domain. Amino acid replacements inT.compared with P. dabryanus (PD) marked with red box.The red star marks theamino acid replacement that affects protein function as predicted by PROVEAN.(B) Amaximum-likelihood tree and alignment of fish pVHL sequences. Aminoacids unique to the YC are highlighted in red, which identified by branch-sitemodel with a posterior probability greater than 0.5. FIGURE S5| The effects of pVHL overexpression in HEK 293T cell on thetranscriptional activity of (A) HIF-as in T. scleroptera (YC), and (B) P.dabryanus(PD) measured with luciferase activity assays. *p<0.05,**p<0.01,***p <0.001, ns represents no significant difference. Error bars indicate thestandard error of the mean. FIGURE S6|The effects of pVHL overexpression in HEK 293T cell on the proteinexpression of (a1,a2) HIF-as in T. scleroptera (YC) under normal normoxia andhypoxia conditions (a3,a4) HIF-as in P. dabryanus (PD) under normal normoxiaand hypoxia conditions. HEK293T cells were co-transfected with equal amountsof Myc-tagged HIF-as expression vector along with HA-tagged pVHLexpression vector. TABLE S1 |The PCR primers used to amplify T. scleroptera and P. dabryanus. TABLE S2|Gene specific primers with restriction enzyme sites for full-lengthpVHL and HIF-a isoforms. TABLE S3|Gene information of T. scleroptera and P.dabryanus. TABLE S4|Comparison amino acid sequence similarity of pVHL and HIF-aisoforms between T. scleroptera and other species. TABLE S5|Amino acid variations of pVHL gene in T.scleroptera. TABLE S6List of HIF-a genes in representive invertebrates and vertebrates. Theblack filled pentagram and empty pentagram represent the existency andinexistence of HIF gene in the selected species. TABLE S7|One-ratio model analysis of HIF-as and pVHL gene. TABLE S8| Selective pressure analyses of hypoxia-inducible factor and pVHLusing PAML Branch-Site Model. Cao, Y. B., Chen, X. Q., Wang, S., Wang, Y. X., and Du, J. Z. (2008). Evolutionand regulation of the downstream gene of hypoxia-inducible factor-lalpha innaked carp (Gymnocypris przewalskii) from Lake Qinghai, China. J. Mol. Evol.67,570-580. doi: 10.1007/s00239-008-9175-4 Cheviron, Z. A., and Brumfield, R. T. (2012). Genomic insights into adaptation tohigh-altitude environments. Heredity 108, 354-361. doi: 10.1038/hdy.2011.85 Choi, Y., Sims, G. E., Murphy, S., Miller, J. R., and Chan, A. P. (2012). Predictingthe functional effect of amino acid substitutions and indels. PLoS ONE 7:e46688.doi: 10.1371/journal.pone.0046688 Cockman, M. E., Masson, N., Mole, D.R., Jaakkola, P.,Chang, G. W., Clifford, S. C.,et al. (2000). Hypoxia inducible factor-alpha binding and ubiquitylation by thevon Hippel-Lindau tumor suppressor protein. J. Biol. Chem. 275, 25733-25741.doi: 10.1074/jbc.M002740200 Edgar,R. C.(2004).MUSCLE: multiple sequence alignment with high accuracy andhigh throughput.Nucleic Acids Res. 32,1792-1797. doi: 10.1093/nar/gkh340 Epstein, A. C. R., Gleadle, J. M., McNeill, L. A., Hewitson, K. S., O’Rourke, J.,Mole, D. R., et al. (2001). C-elegans EGL-9 and mammalian homologs definea family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell 107,43-54.doi: 10.1016/S0092-8674(01)00507-4 Gou, X., Wang, Z., Li, N., Qiu, F., Xu, Z., Yan, D., et al. (2014). Whole-genomesequencing of six dog breeds from continuous altitudes reveals adaptation tohigh-altitude hypoxia. Genome Res. 24, 1308-1315. doi: 10.1101/gr.171876.113 ( Graham, A . M . , and M c Cracken, K. G. (20 1 9). Co n vergent evo l ution on the hypoxia-inducible factor (HIF) pathway genes E GLN1 an d EPAS 1 in h ig h - altitude ducks. Heredity 122 , 819-832. doi: 10.1038/s41437-018-0173-z ) ( G uan, L . , Ch i , W . , X i a o, W., Ch e n, L., a n d He, S. ( 2 014). Anal y sis of h y po x ia- inducible f actor alpha p olyploidization reveals a daptation to T i betan P l ateauin the evolution o f schizothoracine fish. BMC Evol. B i ol.14:192. doi: 1 0 .1186/ s12862-014-0192- 1 ) ( H erzenstein, S. M. ( 1888).“Fische,in Wissenschaftliche Re s ultate der von N. M. Przewalski nach Central-Asien unternommenen Reisen. Zoologischer Theil (St. Petersburg: Kaiserlichen Akademie der Wissenschaften), 54-57. ) ( Hu, C . J , W ang, L . Y . , Chodosh, L. A . , K e ith, B., a nd Simon, M . C. (2 0 03). D ifferential roles of hypoxia-inducible facto r lalpha (HIF-1alpha) and HIF-2alpha in hypoxic gene regulation. Mol. Cell. Biol. 23, 9361-9374. ) ( Huelsenbeck, J. P., and Ronquist, F. (2001). MRBAYES: b a yesian i n ference of phylogenetic trees. Bioinformatics 17, 754-755. d o i: 1 0 .1093/bioinformatics/17.8.754 ) ( I van, M., K ondo, K . , Y a ng, H . , K i m, W. , Valiando, J., Ohh, M. , et al. (2001). H I Falpha targeted for VHL-mediated des t ruction by p roline hyd r oxylation: implications for O2 sensing. Science 292, 464-468. do i : 10 . 1126/science.1059817 ) ( J aakkola, P ., Mole, D . R., T i a n, Y. M., W i lson, M. I, G ie lbert, J. , Gaskell, S. J . ,et al. (2001) . Targeting of HIF-alpha to the von Hippel-Lindau u b iquitylationcomplex by O2-regulated pr o lyl hydroxylation. Sc i ence 292, 468-472. doi: 10. 1126/science.1059796 ) ( Kaelin, W . G . J r ., and R a tcliffe, P . J . (2008). Oxygen se n sing by metazoans: the central role of the HIF hydroxylase pathway. Mol. Cell. 3 0 , 393-402. doi: 1 0 . 1016/j.molcel.2008.04.009 ) ( Kang, J . , Ma, X ., a nd H e , S. ( 2 017). E v idence of high-altitude adaptation inthe glyptosternoid fish, C reteuchiloglanis macropterus fr o m the Nuj i ang Riverobtained t h rough tr a nscriptome analysis. BMC Evol Biol. 17:2 2 9. doi: 10.1 1 86/ s12862-017-1074-0 ) ( Kietzmann, T ., M ennerich, D ., a nd Dimova, E . Y . ( 2 016). H y poxia-induciblefactors (HIFs) and phosphorylation: imp a ct on s t abi l ity, loca l ization, and transactivity. F ront. C ell Dev. Biol. 4 :11. doi: 1 0.3389/fcell.2016.00011 ) ( Koh, M. Y . , a nd P o wis, G. ( 2 012). Passing the b aton: t he H IF switch. T rends Biochem. S c i. 3 7, 364-372. doi: 1 0.1016/j.tibs.2012.06.004 ) ( Lando, D ., P ongratz, I., P oellinger, L. , and Whitelaw, M. L . ( 2000). A r e dox m echanism controls differential D N A bin d ing activities of hypoxia-inducible f actor (HIF) l alpha and t h e H I F-like factor. J . Biol . Chem. 2 75, 4 618-4627. doi: 10.1074/jbc.275.7.4618 ) ( Letunic, I., Doerks, T . , and Bork, P. (2015). SMART: r e cent u p dates, n e wdevelopments and status in 2015. Nu c leic Acids Res. 43, D257-D260. doi: 10.1093/nar/gku949 ) ( Li, J . -T. , Gao, Y.-D., Xie, L., D eng, C . , Shi, P., Guan, M.-L., et al.(2018). Comparative genomic investigation o f h igh- e levation adaptation inectothermic s n akes. P r oc. Natl. Acad. S c i. U .S.A. 11 5 , 8406-8411. doi: 10 . 107 3 / pnas.1805348115 ) ( Liu, X., Zhang, Y., Li, Y ., Pan, J., Wang, D., Chen, W., et a l. (2 0 19). EPAS1 gain-of-function mutation contributes to high-altitude adaptation in Tibetan horses.Mol. Biol. Evol. doi: 10.1093/molbev/msz158 [Ep u b ahead of print]. ) ( Ma, X., Dai, W.,Kang, J., Yang, L., and He, S. (2015). Comprehensive transcriptomeanalysis of six catfish species from a n a l titude gradient r e veals a d aptiveevolution i n t i betan f i shes. G 3 (Bethesda) 6, 141-148. doi: 10.1534/g3.115.024448 ) ( Maynard,M. A . ,and Ohh,M. (2004).von Hippel-Lind a u tumor suppressor proteinand hypoxia-inducible factor i n kidney cancer. Am. J. N e phrol. 24,1-13. doi:10.1159/000075346 ) ( Meyer, A ., a nd Van d e Peer, Y. ( 2 005). From 2R to 3R: evidence for a fish - specific genome duplication (FSGD). Bioessays 27, 9 3 7-945. doi: 1 0 .1002/bies. 20293 ) ( Miao, B., Wang,Z., and Li,Y. (2016). Genomic analysis reveals hypoxia adaptationin the Tibetan mastiff by introgression of the gray wolf from the Tibetan Plateau. Mol. Biol. Evol. 34,734-743. ) ( Nilsson, G. E., and Ostlund-Nilsson, S. (2004). Hypoxia in paradise: widespread hypoxia tolerance in coral reef fis h es. Pr o c. Bi o l. Sci. 27 1 (Su p pl.3), S30- S 33. d oi: 10.1098/rsbl.2003.0087 ) ( Ohno, S . ( 1 971). E v olution b y Ge n e Dup l ication. Be r l in: Spr i nger Sci e nce & Business Media. ) Okuno, H., Akahori, A., Sato, H., Xanthoudakis, S., Curran, T., and Iba, H.(1993). Escape from redox regulation enhances the transforming activity of fos.Oncogene 8, 695-701. Opazo, J. C., Butts, G. T., Nery, M. F., Storz, J. F., and Hoffmann, F. G. (2013).Whole-genome duplication and the functional diversification of teleost fishhemoglobins.Mol. Biol. Evol. 30, 140-153. doi: 10.1093/molbev/mss212 ( Ouyang, M., Yu, X . , a nd Chen, D. Y . ( 2 001). The preliminary studies on oxygen consumption rate and a s phyxia point ofMegalobrama amblycephalain Poyang L ake.J. Jiangxi Fish. Sci. T echnol. 4, 20-22. ) ( Posada,D . (2008). jModelTest: phylogenetic model averaging. Mol. Biol. Evol. 25, 1 253-1 2 56. doi: 10.1093/molbev/msn083 ) ( Prabhakar, N. R ., a nd S emenza, G . L. (2 0 12). Ad a ptive an d mal a daptivecardiorespiratory responses t o continuous and intermittent h y poxia me d iatedby h ypoxia-i n ducible factors 1 a n d 2. Physiol. R e v. 92,967-1003. doi: 10.1152/ physrev.00030.2011 ) ( Qi, D ., C hao, Y . , G u o, S. , Z h ao, L. , Li , T . , W e i, F . , et a l . ( 2 012). C o nvergent, p arallel and correlated evol u tion of t rophic m orphologies i n t h e s u bfamilyschizothoracinae from the Qinghai-Tibetan plateau. PL o S ONE 7:e34070. doi:10.1371/journal.pone.0034070 ) ( R ahman, M . S . , a n d Th o mas, P. ( 2 007). Mol e cular clon i ng, characterization and e xpression o f t wo h ypoxia-inducible f actor alpha subunits, HIF-1alphaand H IF-2alpha, i n a h y poxia-tolerant m arine t e leost, A tlantic croaker (Micropogonias u n dulatus). G e ne 39 6 , 27 3 -282. doi : 10.1016/j.gene.2007.03. 009 ) ( R amirez, J . M., F olkow, L . P., and Blix, A. S. (2007). Hypoxia tolerance in mammalsand b irds: from t he w ilderness to the c linic. Annu. Rev. P hysiol. 69, 113 - 143. doi: 1 0.1146/annurev.physiol.69.031905.163111 ) ( R aval, R . R ., L au, K. W . , Tran, M. G., So w t er, H. M., M a ndr i ota, S. J., L i, J. L., et al.(2005). Contrasting properties of hypoxia-inducible f actor 1 (HIF-1) and H I F-2 in von H i ppel-lindau-associated renal cell carcinoma. Mol. Cell. Bi o l. 2 5 , 5675-5686.doi: 1 0.1128/MCB.25.13.5675-5686.2005 ) ( R ytkonen, K . T . , Akbarzadeh, A. , Miandare, H. K ., Kamei, H., Dua n , C., L ede r , E . H ., et al. (2013). Subfunctionalization of cyprinid hy p oxia-inducible factorsfor roles in development and o x ygen se n sing. Evolution 67 , 873-882. doi: 10 . 1111/j.1558-5646. 2 012.01820.x ) ( Rytkonen, K . T ., Vuori, K. A., Primmer, C. R ., and N ikinmaa, M . ( 2 007). Comparison o f hypoxia-inducible fa c tor-1 a l pha i n hypoxia-sensitive a n dhypoxia-tolerant fish species . Compar. Biochem. Physiol. Part D Genomics P roteomics 2, 177-186. doi: 10.1016/j.cbd.2007.03.001 ) ( Scheinfeldt, L . B . , an d Tishkoff, S. A. (20 1 0). Liv i ng the high l ife: h igh- a ltitude a daptation. Genome Biol. 11:133. doi : 10.1186/gb-2010 -1 1-9-133 ) ( Schofield, C. J ., and Ratcliffe, P. J . (2004) . Oxygen s ensing by HIF hy d roxylases. Nat. Rev. Mol. C ell Biol . 5, 343-354. doi : 10.1038/nrm1366 ) ( Schrodinger, L. L. C. (2010).The PyMOL Molecular Graphics System, Ver s ion 1.3r1.Available online at: http://www.pymol . org ) ( Semenza, G . L . ( 1999). Regulation of mammalian O 2 homeostasis by hypoxia- inducible factor 1 . Annu. R ev. C ell Dev. Biol. 1 5,551-578. d oi: 10. 1 146/annurev. cellbio.15.1.551 ) ( Semenza, G. L. (2012). Hypoxia-inducible factors in physiology and medicine. Cell148, 399-408. d o i: 10.1016/j.cell.2012.01.021 ) ( Shams, I., Avivi, A., and Nevo, E . (2004). Hypoxic stres to l erance of the bl i ndsubterranean m ole rat: expression of erythropoietin and hypoxia-inducible f actor 1 a lpha. P roc. N atl. Acad. Sci. U .S.A. 1 01, 9698-9703. doi: 10 . 1 073/pnas. 0403540101 ) ( Soitamo, A . J ., Rabergh, C. M., Gassmann, M., S i stonen, L ., and N ikinmaa,M.(2001). C h aracterization of a hypoxia-inducible factor (HIF-1alpha) from rainbow t rout-accumulation of protein occurs at normal v enous oxygen t ension. J. Bio l . Chem.276, 19699-19705. doi : 10.1074/jbc.M009057200 ) ( Stantic, M., Wolfsberger, J., Sakil , H . A . M. , and Wilhelm,M . T. ( 2 018).De l taNp73enhances HIF-1alpha protein stability through repression of the ECV c o mplex.Oncogene 37, 3729-3739. doi: 1 0.1038/s41388-018-0195-2 ) ( Stebbins, C. E.,Kaelin, W. G. Jr., and Pavletich,N. P. (1 9 99). Structure of the VHL - E longinC-ElonginB complex: implications for VHL tumor suppressor function. Science 284, 455-461. doi: 10.1126/science.284.5413.455 ) ( Su, J . , and Wang, Z. (1992). S t udies on th e population energetics of plateau zokor: average daily metabolic rate and burrowing metabolic rate. A c ta TheriolSin 12,200-206. ) ( T arade, D., Lee, J. E., an d Ohh, M. (2019). Evolution of metazoan o x ygen-sensing involved a conserved divergence of VHL affinity fo r HIF1o and HIF2a. Nat. Commun. 10,3293. doi: 1 0.1038/s41467-019-11149-1 ) ( Terova, G., R imoldi, S . , Cora, S . , Bernardini, G . , Gornati, R., a n d Sa r oglia, M.(2008). Acute and chronic hypoxia affects HIF-1o mRNA levels in sea bas s (Dicentrarchus l a brax). A q uaculture 279, 150 - 159 . doi: 10.1 0 16/j.aquaculture. 2008.03.041 ) ( Tian,R., W ang, Z., Niu, X., Zhou, K., Xu, S ., and Yang, G. (2016). Evolutionary g enetics of hypoxia t o lerance in c e taceans du r ing diving. Genome Biol. Evol . 8, 827-839. doi: 10.1093/gbe/evw037 ) ( Wang, G. L., Jiang, B. H., Rue, E. A., and Semenza, G. L . ( 1995). Hypoxia-inducible factor 1 i s a basic-helix-loop- h elix- P AS h e terodimer reg u lated by cellular O2tension. Proc. Natl. Acad. S ci. U.S.A. 92, 5510-5514. doi: 10.1073/pnas.92.12.5510 ) ( Wang, G. L., a nd S emenza, G. L. ( 1993). General Involvement of Hypoxia-Inducible Factor-I i n t r anscriptional response to hypoxia. Proc. Natl. Acad.Sci. U.S.A. 90, 4304-4308. doi: 10.1073/pnas.90.9.4304 ) ( Wang, Y . , S h en, Y. , Fe n g, C. , Zh a o, K., Son g , Z., Zha n g, Y., et a l . ( 2016). M itogenomic perspectives on the origin of Tibetan loaches and the i r adaptation t o high altitude. Sci Rep 6:29690. doi: 10.1038/srep29690 ) ( Wang, Y., Yang,L., Wu, B., Song, Z., and He, S. ( 2 015a).Transcriptome analysis of t he p lateau fish ( Triplophysa dalaica): implications for adaptation to hypoxia in fishes. Gene 565, 211-220.doi: 10.1016/j.gene.2015.04.023 ) ( Wang, Y., Yang, L., Zhou, K ., Zhang,Y., S ong, Z., and He,S. ( 2015b). E vidence foradaptation to the tibetan plateau inferred from tibetan loach transcriptomes. Genome Biol. E vol. 7,2970- 2 982. doi:10.1093/gbe/evv192 ) ( Wang, Z., Yonezawa, T., L iu , B., Ma, T., Shen, X., Su, J., et al.(2011). Domestication relaxed selective constraints on the yak mitochondrial g enome. M o l. Biol. Ev o l.28,1553-1556.doi: 10.1093/molbev/msq336 ) ( Webb, J . D., Coleman, M. L ., and Pugh, C. W.(2009). Hypoxia, hypoxia- i nduciblefactors (HIF), H IF h y droxylases an d oxygen sens i ng. Cell Mol. Life . Sci 66,3539-3554. doi: 10.1007/s00018-009-0147-7 ) ( Wu, Y . , and W u , C. (1992). Th e Fishes of the Qinghai-Xizang Plateau. Chengdu: Sichuan Science and Technology Press. ) ( Xu, Q., Zhang, C.,Zhang, D.,Jiang, H., Peng,S.,Liu, Y., et al.(2016). Ana l ysis ofthe erythropoietin of a Tibetan Plateau schizothoracine fi s h (Gymnocypris dobula) ) reveals enhanced cytoprotection function in hypoxic environments. BMC Evol.Biol. 16:11. doi: 10.1186/s12862-015-0581-0 Xu, S., Li, S., Yang, Y., Tan, J., Lou, H., Jin, W., et al. (2011). A genome-widesearch for signals of high-altitude adaptation in Tibetans. Mol. Biol. Evol. 28,1003-1011. doi: 10.1093/molbev/msq277 Yang, L., Wang, Y., Zhang, Z., and He, S. (2014). Comprehensive transcriptomeanalysis reveals accelerated genic evolution in a Tibet fish, Gymnodiptychuspachycheilus.Genome Biol. Evol. 7, 251-261. doi: 10.1093/gbe/evu279 ( Yang, Z ., Wong, W. S . , and N i elsen, R. (2005). B ayes empirical bayes inference of amino acid sites u nder positive s election. Mol. Biol. Evol . 22 , 1107-1118. doi: 10.1093/molbev/msi097 ) Yang, Z. H. (2007). PAML 4: phylogenetic analysis by maximum likelihood.Mol.Biol.Evol.24, 1586-1591. doi: 10.1093/molbev/msm088 Zhang, D., Yu, M., Hu, P., Peng, S., Liu, Y., Li, W., et al. (2017). Genetic adaptationof schizothoracine fish to the phased uplifting of the qinghai-tibetan plateau. G3(Bethesda) 7,1267-1276. doi: 10.1534/g3.116.038406 ( Zhang, W ., F a n, Z . , H a n, E. , H o u, R., Zh a ng, L., Galaverni, M. , e t al. (2014). Hypoxia adaptations in the grey wolf (Canis lupus chanco) fromQinghai-T i bet Plateau. P L oS Genet. 10 : e1004466. doi: 10.1371/journal.pgen.100 4 466 ) ( Zhou, S. Z ., Wang, X. L . , Wang, J., and Xu, L. B. ( 2006). A p reliminary study o n timing of the oldest Pleistocene glaciation i n Qinghai-Tibetan P lateau. Quat. Int. 154,44-5 1 . doi: 10.1016/j.quaint.2006.02.002 ) ( Conflict o f Interest: The authors declare that t h e research was co n ducted in the absence of any commercial or fi n ancial relationships that could b e c o nstrued as apotential conflict of interest. ) ( Copyright 2 020 Chen, Shen, Wang, Ouyang, Kang, Ly, Ya n g and He. Thi s is anopen-access article distributed under the terms of the Creative Commons AttributionLicense ( CC BY). T h e use, distribution o r r e production in other forums is permitted, provided the original author(s) and th e copyright ow n er(s) are cre d ited and that the original publication in this journal i s cited, i n a ccordance wi t h accepted academicpractice. No use, distribution or reproduction is permitted which d oes not comply with these terms. ) Frontiers in Geneticswww.frontiersin.orgMay Volume Article 文章题目:Analysis of Multiplicity of Hypoxia-Inducible Factors in the Evolution of Triplophysa Fish (Osteichthyes: Nemacheilinae) Reveals Hypoxic Environments Adaptation to Tibetan Plateau三鳃鱼缺氧诱导因子的多样性分析揭示了青藏高原低氧环境的适应文章出处:Front Genet, 2020, 11: 433. 中国科学院水生生物研究所,武汉工作站使用情况:InvivO2 400使用气体浓度:低氧(2% O2)摘要:HIF(缺氧诱导因子)基因家族成员在氧利用率变化期间作为细胞和全身氧稳态的主要调节者。青藏高原是形成长期缺氧和冷适应天然实验室。在这种情况下,限于3500 m 高海拔淡水河流的硬鳍石斑鱼被选为模型,以与平原的代表性物种大鳞石斑鱼进行比较。我们克隆了不同的HIF-α,并进行从无脊椎动物到脊椎动物的系统发育分析,以鉴定HIF-α基因并分析其进化史。有趣的是,HIF-α经历了基因复制,这可能是由于进化过程中的全基因组复制(WGD)事件。PAML(发育分析软件)分析表明HIF-1αA 在三鳃鱼谱系中受到正向选择作用;为了研究高原和平原鱼类低氧适应与肿瘤抑制蛋白pVHL 调节HIF-α稳定性的关系,进行了一系列实验。比较HIF-α的荧光素酶转录活性和蛋白水平以及HIF-αs 与pVHL 的不同相互作用,显示了高原鱼和平原鱼之间的明显差异。通过pVHL 蛋白突变进行的功能验证表明,这些突变增加了HIF-α的稳定性及其对芳香烃受体核转位器ARNT 的异源二聚化亲和力。我们的研究表明,pVHL 的错义突变诱导了生活在高海拔低氧环境中的高原鳅的进化分子适应。FIGURE 1 | Phylogenetic tree of the HIF-α genes. The maximum likelihood method was used to reconstruct the tree that was rooted with invertebrate HIF-α sequences. Species are listed along with their phylums or classes.FIGURE 4 | Transcriptional activity analysis of different HIF-αs under normoxic and hypoxic conditions. (A) Multiple sequence alignment of the N-terminus of the HIF-2α bHLH domain amino acid sequences. The basic domain that interacts directly with DNA is marked with a solid dash line and the variation between serine and cysteine at position 28 is marked by a regular red triangle. YC, T. scleroptera, PD: P. dabryanus TD, T. dalaica; TS, T. siluroides. (B) Transcriptional activity comparative analysis of different HIF-αs between high-elevation T. scleroptera (YC) and low-elecation P.dabryanus (PD). Significant differences in the transcriptional activity of HIF-2αA. Error bars represent the mean ± SEM. Statistical significance was determined using the two-tailed unpaired Student t-test with P< 0.05. HIF-α基因的系统进化树(图1);脊索动物亚门底部的文昌鱼只有一个HIF-α基因,而脊椎动物通常有两个或多个来自两个全基因组复制事件的旁系同源物,这表明文昌鱼是HIF-α家族的古老起源;另外两种旁系同源物(HIF-1αA/B 和HIF-2αA/B)是存在于鲤科鱼类中,很可能是硬骨鱼类特有的结果; 在常氧和低氧条件下不同HIF-α的转录活性分析(图4);低氧可以增强HIF-α的转录活性,与平原泥鳅相比,高原泥鳅的HIF-2αA 无论在常氧还是低氧条件下都表现出更强的转录活性,在其他HIF-α旁系同源物之间没有显著差异(图4B)。
确定










还剩13页未读,是否继续阅读?
北京隆福佳生物科技有限公司为您提供《三鳃鱼中HIF(缺氧诱导因子)多样性分析检测方案(其它培养箱)》,该方案主要用于其他中种类检测,参考标准--,《三鳃鱼中HIF(缺氧诱导因子)多样性分析检测方案(其它培养箱)》用到的仪器有









