DNA fragments ranging from 25 to 2000 base pairs (bp) in length were used for the
development of a suitable gradient for separating DNA molecules
方案详情

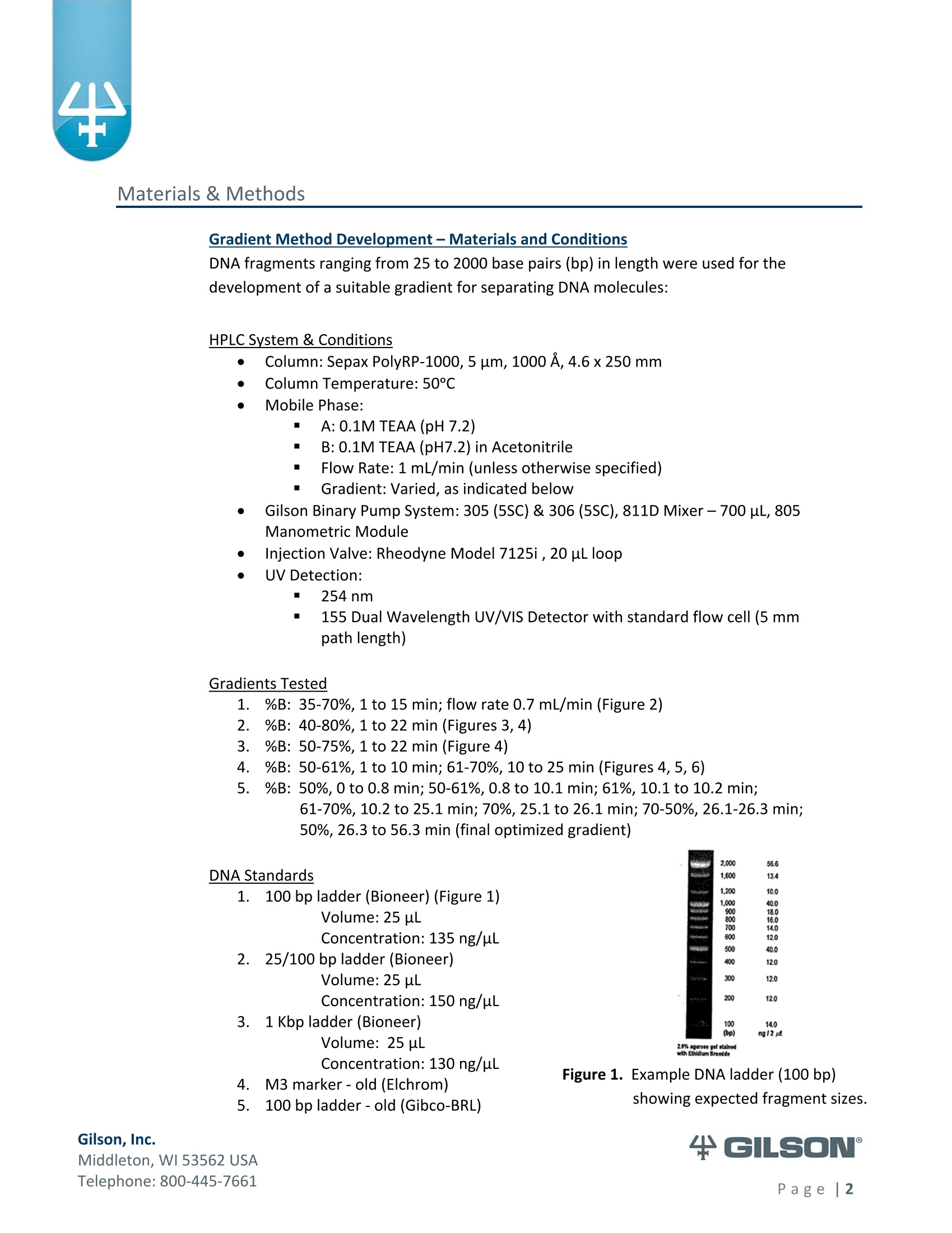
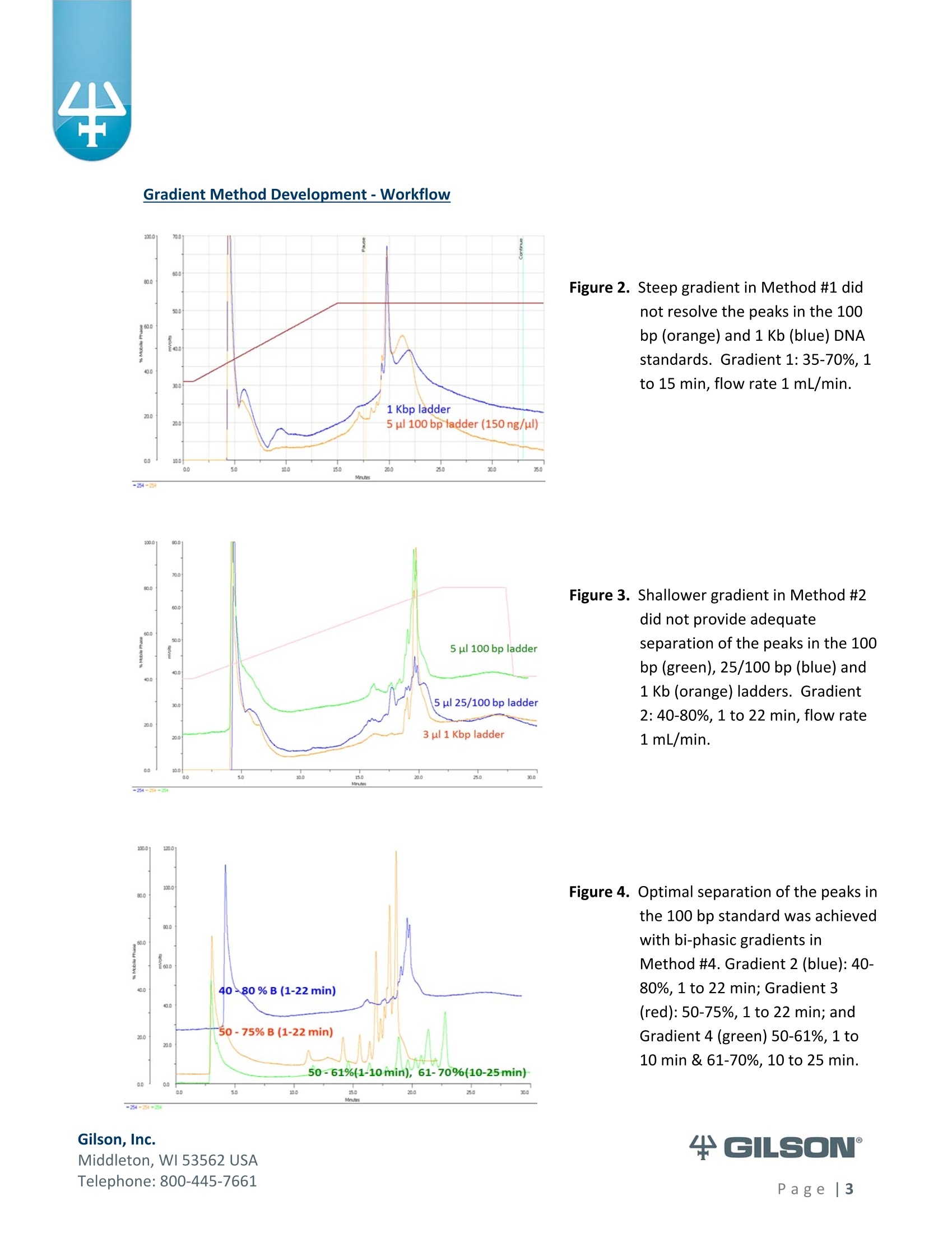
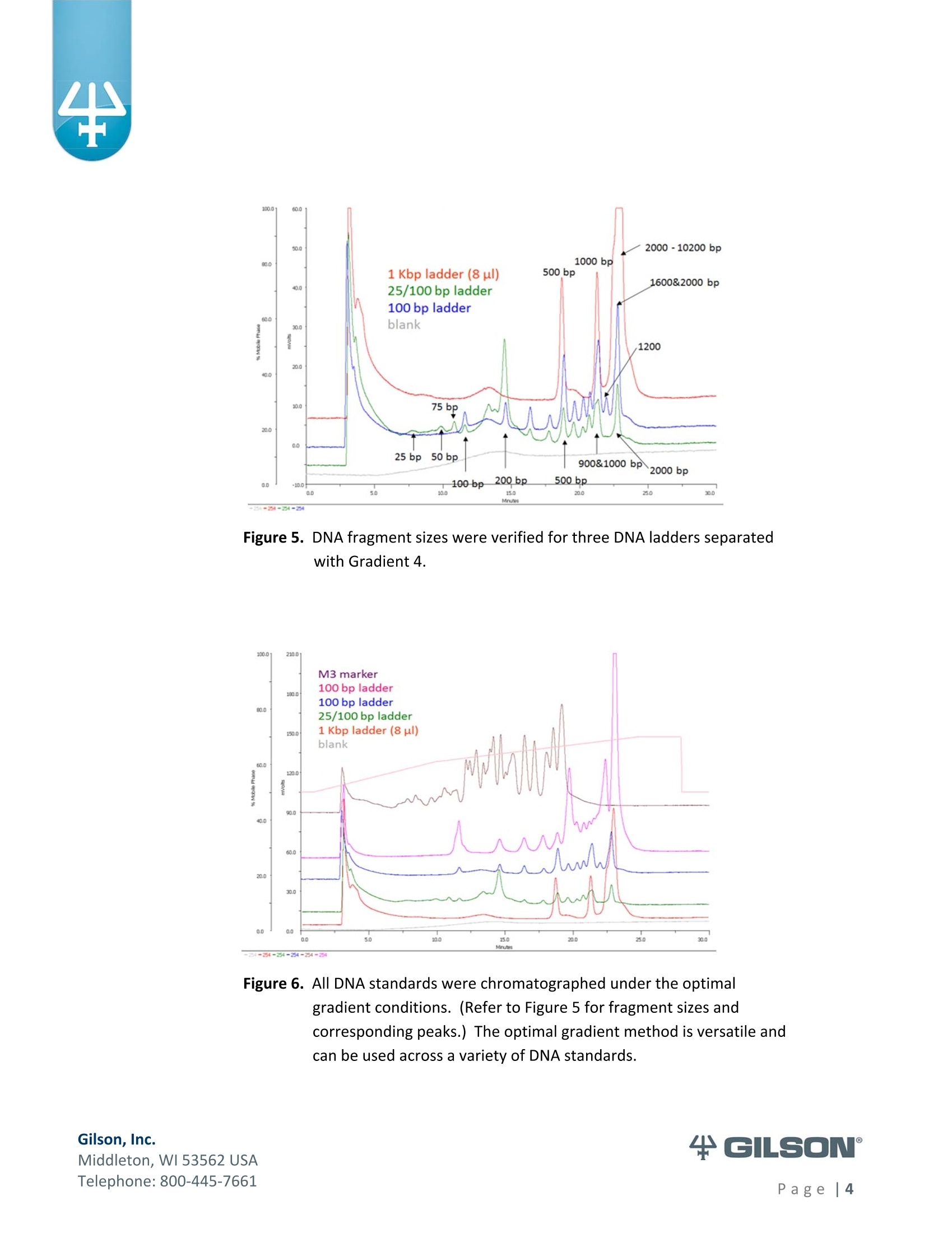
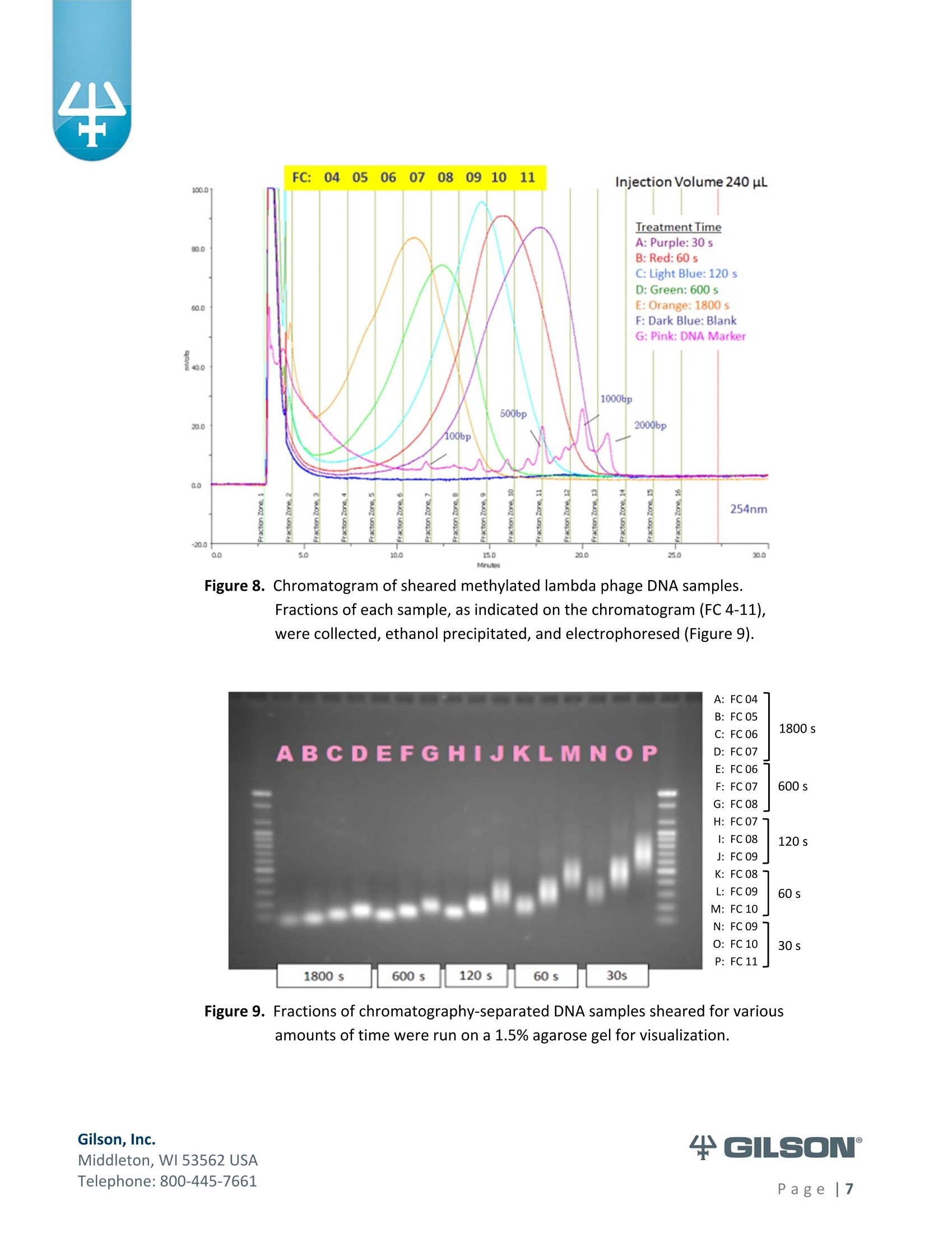
Materials & Methods Results High-Resolution Purification of Sheared LambdaPhage DNA Using a Gilson HPLC System Application Note CL0513 Keywords High-Performance Liquid Chromatography (HPLC), DNA shearing, DNA separation,methylation,lambda phage,gradient pattern, electrophoresis,next-generationsequencing (NGS) Introduction Data in this application note were provided by M. Kawakatsu and Y. Hayashi, AppliedTechnical Department, M&S Instruments, Inc., Japan. As the size-selective separation of nucleic acids is a necessary step in many molecularbiological analyses performed in clinical, forensic, and other types of laboratories, thedevelopment of optimal DNA separation methods is important. Liquidchromatographic separation techniques involve simple automated sample purificationsteps and provide high resolution of DNA molecules, thereby enabling efficient DNAseparation. Furthermore, fragments can be identified and quantified directly whenusing liquid chromatography methods. The separation and characterization of modified DNA is particularly relevant, asgeneral interest in next-generation sequencing (NGS) is expanding. This applicationnote describes the successful separation of methylated lambda phage DNA fragmentson a Gilson HPLC system using a Sepax PolyRP-1000 column. Prior to HPLC analysis, agradient method was developed to provide sufficient resolution of the DNA fragments(25-2000 bp). These results demonstrate the feasibility of separating DNA fragments,including those containing methylated modifications, by HPLC. This highly effectiveseparation method can thus be applied in a wide variety of analyses, including thepreparation steps of next-generation sequencing workflows. Gradient Method Development-Materials and Conditions DNA fragments ranging from 25 to 2000 base pairs (bp) in length were used for thedevelopment of a suitable gradient for separating DNA molecules: HPLC System & Conditions Column: Sepax PolyRP-1000, 5 um, 1000 A, 4.6x250mm Column Temperature:50°℃ Mobile Phase: A: 0.1M TEAA (pH 7.2) B: 0.1M TEAA (pH7.2) in Acetonitrile Flow Rate: 1 mL/min (unless otherwise specified) Gradient:Varied, as indicated below Gilson Binary Pump System: 305 (5SC) & 306 (5SC), 811D Mixer-700 pL, 805Manometric Module Injection Valve: Rheodyne Model 7125i , 20 uLloop UV Detection: 254nm m 155 Dual Wavelength UV/VIS Detector with standard flow cell (5 mmpath length) Gradients Tested 1%B: 35-70%, 1 to 15 min; flow rate 0.7 mL/min (Figure 2) %B: 40-80%, 1 to 22 min (Figures 3,4) %B: 50-75%, 1 to 22 min (Figure 4) 4%B: 50-61%, 1 to 10 min; 61-70%, 10 to 25 min (Figures 4, 5, 6)5%B: 50%,0 to 0.8 min; 50-61%, 0.8 to 10.1 min; 61%, 10.1 to 10.2 min; 61-70%, 10.2 to 25.1 min; 70%, 25.1 to 26.1 min; 70-50%,26.1-26.3min;50%, 26.3 to 56.3 min (final optimized gradient) 2.000 56.6 DNA Standards 1.60( 13.4 1,200 10 18.0 09 400 120 300 12.0 200 12.0 14.0o ng12 ul 2.0% agaroso gel stainieed with Ethidhum Bromlde 4 M3 marker-old (Elchrom) 5. .1100 bp ladder-old (Gibco-BRL) Figure 1. Example DNA ladder (100 bp)showing expected fragment sizes. Gradient Method Development -Workflow -254 Gilson, Inc. Middleton, WI 53562 USA Telephone: 800-445-7661 Figure 2. Steep gradient in Method #1 didnot resolve the peaks in the 100bp(orange) and 1 Kb (blue) DNAstandards. Gradient 1:35-70%, 1to 15 min, flow rate 1 mL/min. Figure 3. Shallower gradient in Method #2did not provide adequateseparation of the peaks in the 100bp (green), 25/100 bp (blue) and1 Kb (orange) ladders. Gradient2: 40-80%, 1 to 22 min, flow rate1 mL/min. Figure 4. Optimal separation of the peaks in the 100 bp standard was achievedwith bi-phasic gradients inMethod #4. Gradient 2 (blue):40-80%, 1 to 22 min; Gradient 3(red): 50-75%, 1 to 22 min; andGradient 4 (green) 50-61%, 1 to10 min & 61-70%, 10 to 25 min. Figure 5. DNA fragment sizes were verified for three DNA ladders separatedwith Gradient 4. Figure 6. All DNA standards were chromatographed under the optimalgradient conditions. (Refer to Figure 5 for fragment sizes andcorresponding peaks.) The optimal gradient method is versatile andcan be used across a variety of DNA standards. ( Gilson, I nc. ) ( M i ddl et o n, W I 535 6 2 U S A ) ( T el ep h o ne: 8 0 0 -4 45 - 7 6 61 ) Methods-DNA Shearing and Analysis DNA Shearing DNA Sample: Lambda phage DNA, methylated (Sigma-Aldrich) 7 ug in 300 pL TE Buffer (pH 8.0) Shearing Conditions: Covaris S2 Focused-ultrasonicator (Covaris, Inc.) ■ Duty Cycle-20%; Intensity-8; Cycle/Burst-200 Treatment times: 30 s, 60 s, 120 s, 600 s, 1800 s HPLC System and Conditions Column: Sepax PolyRP-1000,5 um, 1000 A, 4.6x250 mm Column Temperature: 50°℃ Mobile Phase: A: 0.1M TEAA (pH 7.0) B: 0.1M TEAA (pH7.0)+25% Acetonitrile Gradient 5: 0-0.8 min (50%), 0.8-10.1 min (50-61%), 10.1-10.2 min (61%), 10.2-25.1 min (61-70%), 25.1-26.1 min (70%), 26.1-26.3 min (70-50%), 26.3-56.3min(50%) Gilson Binary Pump System: 305 (5SC), 306 (5SC), 811D Mixer-700 pL, 805Manometric Module Injection Valve: Rheodyne Model 7725i, 500 uL loop UV Detection: 254nm 155 Dual Wavelength UV/VIS Detector with standard flow cell (5 mm pathlength) Fraction Concentration Method: Ethanol precipitation Electrophoresis and Imaging 1.5% agarose gel using the AgaroPower kit (Bioneer) E-Box used for gel imaging (Vilber-Lourmat) Telephone: 800-445-7661 Results produced by the chromatographic separation of sheared DNA samples usingoptimized gradient conditions were consistent with the standard electrophoresisseparation method, as indicated below in Figures 7, 8, and 9. Reproducibility of the Covaris S2 DNA shearing method was first confirmed bystandard gel electrophoresis, wherein two independent sets of sheared DNAsamples were separated and compared (Figure 7). Figure 7. Sheared DNA samples separated by agarose gel electrophoresis. The tworeplicates showed comparable fragment lengths, indicating adequatereproducibility. When the Gilson HPLC system combined with a Sepax PolyRP-1000 column wasemployed as an alternative separation method, the separation of sheared DNAfragments was similar to that detected by standard gel electrophoresis. Using thegradient conditions described (p. 5), adequate fragment separation was achieved.Moreover, when HPLC fractions containing the different fragment sizes (Figure 8)were run on an agarose gel and visualized, apparent fragment resolution wasconsiderably higher (Figure 9) than fragments separated without HPLC. ( T el ep hon e : 8 00 - 4 45 - 7 6 61 ) Figure 8. Chromatogram of sheared methylated lambda phage DNA samples.Fractions of each sample, as indicated on the chromatogram (FC 4-11),were collected, ethanol precipitated, and electrophoresed (Figure 9). Figure 9. Fractions of chromatography-separated DNA samples sheared for variousamounts of time were run on a 1.5% agarose gel for visualization. ( T el ep h o ne: 8 0 0 -4 45 - 7 6 61 ) Summary High-resolution separation of sheared methylated DNA fragments ranging fromapproximately 25 bp to 2000 bp was readily achieved using HPLC with a Gilsonbinary pump system equipped with a dual wavelength detector. After testingseveral gradient methods, the optimal conditions were defined for an LC system anda Sepax PolyRP-1000 column. Thus, as an accurate and efficient DNA separationtechnique, HPLC represents a feasible, if not better, alternative to traditional gelelectrophoresis methods for DNA sizing. Moreover, the apparent success of thisapproach to separating methylated DNA samples implies its suitability as a methodto follow the DNA shearing step in next-generation sequencing efforts. References 1.Shendure, J., Hanlee J. Next-generation DNA sequencing. Nat.Biotechnol.(2008)26:1135-1145. Gilson, Inc.Middleton, WI USAPage| Gilson, Inc.Middleton, WI USATelephone: age Data in this application note were provided by M. Kawakatsu and Y. Hayashi, AppliedTechnical Department, M&S Instruments, Inc., Japan.As the size‐selective separation of nucleic acids is a necessary step in many molecularbiological analyses performed in clinical, forensic, and other types of laboratories, thedevelopment of optimal DNA separation methods is important. Liquidchromatographic separation techniques involve simple automated sample purificationsteps and provide high resolution of DNA molecules, thereby enabling efficient DNAseparation. Furthermore, fragments can be identified and quantified directly whenusing liquid chromatography methods.The separation and characterization of modified DNA is particularly relevant, asgeneral interest in next‐generation sequencing (NGS) is expanding.1 This applicationnote describes the successful separation of methylated lambda phage DNA fragmentson a Gilson HPLC system using a Sepax PolyRP‐1000 column. Prior to HPLC analysis, agradient method was developed to provide sufficient resolution of the DNA fragments(25‐2000 bp). These results demonstrate the feasibility of separating DNA fragments,including those containing methylated modifications, by HPLC. This highly effectiveseparation method can thus be applied in a wide variety of analyses, including thepreparation steps of next‐generation sequencing workflows.
确定




还剩6页未读,是否继续阅读?
香港华运有限公司为您提供《肠杆菌噬菌体DNA切片中高分辨率纯化检测方案(制备液相色谱)》,该方案主要用于医疗/卫生中高分辨率纯化检测,参考标准--,《肠杆菌噬菌体DNA切片中高分辨率纯化检测方案(制备液相色谱)》用到的仪器有
相关方案
更多









