如何进行长片段基因组DNA质控?—Pippin Pulse脉冲场电泳系统
随着三代测序技术的发展和普及,全基因组测序技术也取得了长足发展。基因组DNA文库的制备质量直接影响测序结果,因而基因组DNA的质控越来越受到重视。而实验室常规的琼脂糖凝胶电泳只能分离20kb以下的DNA分子,当DNA分子大于20kb时,就无法实现有效分离。
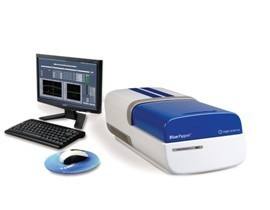
针对超长DNA片段的分离和质控,美国Sage Science公司拥有超高性价比的Pippin Pulse脉冲场电泳系统。Pippin Pulse使用简单,无专用耗材,自带预设程序,能很好的电泳分离0.5Kb到430Kb的基因组DNA片段。Pippin Pulse具有一个简单的软件界面,自带6种预设程序,可以直接分离多种不同大小范围DNA长片段,此外,软件还支持自定义模式,用户可以自编程分离不同范围的DNA片段。Pippin Pulse包括电源、电泳槽、操控仪器的电脑和用户手册,无特殊耗材,使用成本低,经济耐用。

Pippin Pulse脉冲场电泳系统
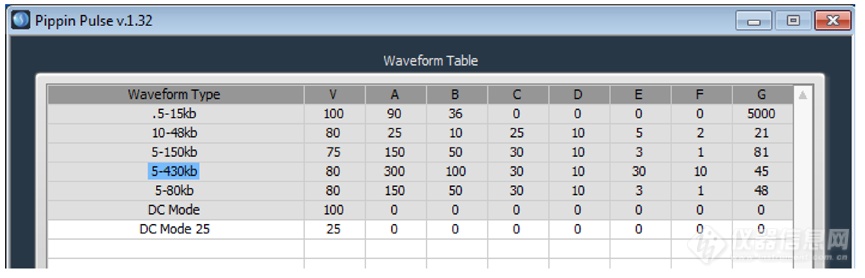
Pippin Pulse脉冲场电泳系统预设程序
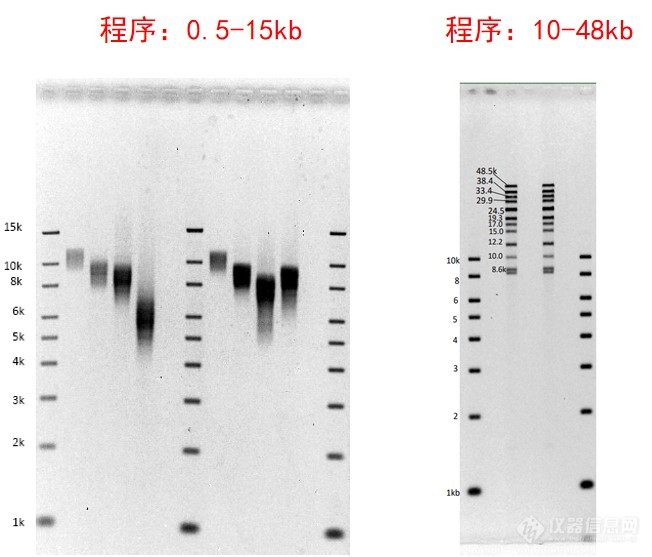
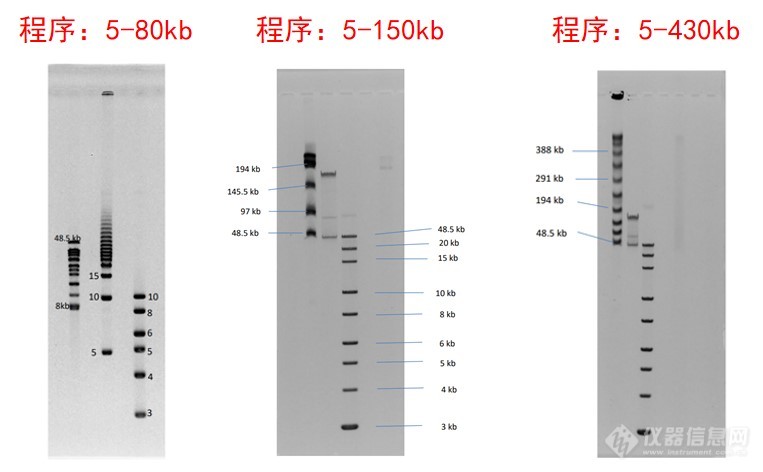
Pippin Pulse脉冲场电泳系统结果图示
Pippin Pulse脉冲场电泳系统应用案例(部分展示)
1. Pippin Pulse 与PacBio三代测序仪联用,控制文库片段分布,充分发挥长片段测序优势
|
|
|
|
|
|
|
|
2. Pippin Pulse 与 Nanopore三代测序仪联用,质控更长、更高质量的DNA,进一步提升测序长度
|
|
|
|
|
|
|
|
3. Pippin Pulse 与 Bionano分子光谱仪联用,确保HMW DNA,保证分析结果准确性
|
|
|
|
|
|
|
|
环亚生物科技有限公司作为美国Sage Science公司Pippin Pulse脉冲场电泳系统在中国区的独家代理商,自2011年以来将Sage Science的产品线引入国内,一直为国内用户提供专业的全自动核酸片段回收系统的安装测试、应用培训,技术支持与售后维护工作,赢得客户的一致好评与信任。环亚生物科技有限公司将一如既往地支持越来越多的Sage Science用户。
▼更多 Pippin Pulse 产品详情,请联系我们▼
环亚生物科技有限公司
地址:上海市闵行区友东路358号闵欣大厦1号楼701/703/705室
电话:021-54583565
邮箱:info@apgbio.com
网址:www.apgbio.com
▼更多精彩内容,请扫码关注我们▼

更多![]()
文献速递|Origins高分辨率潜水电泳仪助力罕见肿瘤患者诊疗 -------微卫星分析
厂商
2024.07.03
Biofluidix 压电式纳升分液装置Pipejet用于透皮给药系统
厂商
2024.07.02
文献速递|全自动DNA片段回收仪Blue Pippin助力废水中病原微生物监测
厂商
2024.07.02
Regemat 3D生物打印神经组织工程材料的性能评估
厂商
2024.07.02